Model info
| Transcription factor | Stat5a | ||||||||
| Model | STA5A_MOUSE.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 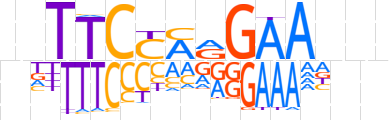 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 14 | ||||||||
| Quality | A | ||||||||
| Consensus | ndnTTCYTRGAAnn | ||||||||
| wAUC | 0.9185652920874283 | ||||||||
| Best AUC | 0.961199934260125 | ||||||||
| Benchmark datasets | 6 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | STAT factors{6.2.1} | ||||||||
| TF subfamily | STAT5A{6.2.1.0.5} | ||||||||
| MGI | 103036 | ||||||||
| EntrezGene | 20850 | ||||||||
| UniProt ID | STA5A_MOUSE | ||||||||
| UniProt AC | P42230 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 28.767 | 20.744 | 39.853 | 18.418 | 76.456 | 10.884 | 7.621 | 48.083 | 41.121 | 19.563 | 46.761 | 17.93 | 36.197 | 13.998 | 42.429 | 31.177 |
| 02 | 48.802 | 24.503 | 50.999 | 58.237 | 13.189 | 15.947 | 3.069 | 32.982 | 34.349 | 37.99 | 23.519 | 40.807 | 11.181 | 30.867 | 25.133 | 48.426 |
| 03 | 1.005 | 0.0 | 0.0 | 106.517 | 0.0 | 1.961 | 0.0 | 107.346 | 0.0 | 0.938 | 0.0 | 101.782 | 0.0 | 0.0 | 0.944 | 179.507 |
| 04 | 0.0 | 0.0 | 1.005 | 0.0 | 0.0 | 0.0 | 0.0 | 2.899 | 0.0 | 0.0 | 0.0 | 0.944 | 32.546 | 7.938 | 5.664 | 449.004 |
| 05 | 0.0 | 31.61 | 0.936 | 0.0 | 2.065 | 5.873 | 0.0 | 0.0 | 0.0 | 6.669 | 0.0 | 0.0 | 1.91 | 447.064 | 0.936 | 2.937 |
| 06 | 0.0 | 2.974 | 1.001 | 0.0 | 16.283 | 237.069 | 14.683 | 223.18 | 0.0 | 0.0 | 0.0 | 1.873 | 0.0 | 2.004 | 0.933 | 0.0 |
| 07 | 0.0 | 0.0 | 10.06 | 6.223 | 0.0 | 0.0 | 22.649 | 219.399 | 0.0 | 0.0 | 5.029 | 11.587 | 0.0 | 0.0 | 157.794 | 67.259 |
| 08 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 80.002 | 1.863 | 105.775 | 7.892 | 54.377 | 0.0 | 245.013 | 5.079 |
| 09 | 0.945 | 0.0 | 132.404 | 1.03 | 0.0 | 0.0 | 1.863 | 0.0 | 1.946 | 0.0 | 343.934 | 4.907 | 0.0 | 0.0 | 12.971 | 0.0 |
| 10 | 2.891 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 450.695 | 0.0 | 14.68 | 25.797 | 4.992 | 0.0 | 0.945 | 0.0 |
| 11 | 453.727 | 0.0 | 0.955 | 3.895 | 0.0 | 0.0 | 0.0 | 0.0 | 15.625 | 0.0 | 0.0 | 0.0 | 25.797 | 0.0 | 0.0 | 0.0 |
| 12 | 189.196 | 115.631 | 80.936 | 109.386 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.955 | 0.96 | 1.93 | 1.006 | 0.0 |
| 13 | 48.975 | 46.652 | 39.806 | 54.722 | 27.207 | 28.33 | 4.902 | 57.121 | 15.643 | 30.545 | 11.995 | 23.759 | 13.584 | 24.287 | 21.224 | 51.247 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.082 | -0.404 | 0.241 | -0.52 | 0.887 | -1.032 | -1.374 | 0.427 | 0.272 | -0.461 | 0.399 | -0.546 | 0.145 | -0.788 | 0.303 | -0.002 |
| 02 | 0.441 | -0.24 | 0.485 | 0.617 | -0.846 | -0.661 | -2.214 | 0.053 | 0.093 | 0.193 | -0.28 | 0.264 | -1.006 | -0.012 | -0.215 | 0.434 |
| 03 | -3.123 | -4.4 | -4.4 | 1.218 | -4.4 | -2.6 | -4.4 | 1.225 | -4.4 | -3.172 | -4.4 | 1.172 | -4.4 | -4.4 | -3.167 | 1.738 |
| 04 | -4.4 | -4.4 | -3.123 | -4.4 | -4.4 | -4.4 | -4.4 | -2.264 | -4.4 | -4.4 | -4.4 | -3.167 | 0.04 | -1.335 | -1.654 | 2.654 |
| 05 | -4.4 | 0.011 | -3.173 | -4.4 | -2.557 | -1.62 | -4.4 | -4.4 | -4.4 | -1.5 | -4.4 | -4.4 | -2.622 | 2.649 | -3.173 | -2.253 |
| 06 | -4.4 | -2.242 | -3.126 | -4.4 | -0.641 | 2.016 | -0.742 | 1.955 | -4.4 | -4.4 | -4.4 | -2.639 | -4.4 | -2.582 | -3.176 | -4.4 |
| 07 | -4.4 | -4.4 | -1.108 | -1.566 | -4.4 | -4.4 | -0.317 | 1.938 | -4.4 | -4.4 | -1.765 | -0.971 | -4.4 | -4.4 | 1.609 | 0.76 |
| 08 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | 0.933 | -2.643 | 1.211 | -1.34 | 0.549 | -4.4 | 2.049 | -1.756 |
| 09 | -3.167 | -4.4 | 1.434 | -3.105 | -4.4 | -4.4 | -2.643 | -4.4 | -2.607 | -4.4 | 2.387 | -1.788 | -4.4 | -4.4 | -0.862 | -4.4 |
| 10 | -2.267 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | 2.657 | -4.4 | -0.742 | -0.189 | -1.772 | -4.4 | -3.166 | -4.4 |
| 11 | 2.664 | -4.4 | -3.159 | -2.0 | -4.4 | -4.4 | -4.4 | -4.4 | -0.681 | -4.4 | -4.4 | -4.4 | -0.189 | -4.4 | -4.4 | -4.4 |
| 12 | 1.79 | 1.299 | 0.944 | 1.244 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -3.159 | -3.156 | -2.614 | -3.122 | -4.4 |
| 13 | 0.445 | 0.397 | 0.239 | 0.555 | -0.137 | -0.097 | -1.788 | 0.598 | -0.68 | -0.023 | -0.938 | -0.27 | -0.817 | -0.249 | -0.381 | 0.49 |