Model info
| Transcription factor | VDR | ||||||||
| Model | VDR_HUMAN.H10MO.B | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 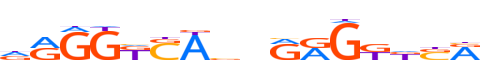 | ||||||||
| LOGO (reverse complement) | 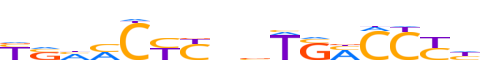 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 16 | ||||||||
| Quality | B | ||||||||
| Consensus | dRGGKSAbnRRGKdSR | ||||||||
| wAUC | 0.8042726613719091 | ||||||||
| Best AUC | 0.902954413484342 | ||||||||
| Benchmark datasets | 4 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | Thyroid hormone receptor-related factors (NR1){2.1.2} | ||||||||
| TF subfamily | Vitamin D receptor (NR1I){2.1.2.4} | ||||||||
| HGNC | 12679 | ||||||||
| EntrezGene | 7421 | ||||||||
| UniProt ID | VDR_HUMAN | ||||||||
| UniProt AC | P11473 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 193.249 | 33.976 | 216.946 | 55.83 |
| 02 | 226.347 | 4.846 | 256.616 | 12.192 |
| 03 | 66.16 | 2.864 | 409.17 | 21.805 |
| 04 | 15.124 | 9.602 | 413.976 | 61.298 |
| 05 | 40.165 | 37.32 | 114.778 | 307.737 |
| 06 | 16.341 | 360.157 | 86.569 | 36.933 |
| 07 | 405.115 | 11.17 | 38.805 | 44.91 |
| 08 | 73.334 | 164.389 | 179.884 | 82.393 |
| 09 | 140.139 | 87.76 | 113.465 | 158.635 |
| 10 | 105.972 | 31.331 | 349.076 | 13.621 |
| 11 | 265.989 | 8.85 | 211.898 | 13.263 |
| 12 | 26.767 | 5.635 | 445.638 | 21.96 |
| 13 | 22.896 | 16.646 | 229.271 | 231.186 |
| 14 | 64.758 | 43.204 | 116.987 | 275.052 |
| 15 | 17.008 | 290.08 | 116.104 | 76.808 |
| 16 | 337.871 | 38.288 | 89.422 | 34.419 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.431 | -1.27 | 0.546 | -0.791 |
| 02 | 0.588 | -2.984 | 0.713 | -2.22 |
| 03 | -0.625 | -3.355 | 1.177 | -1.69 |
| 04 | -2.027 | -2.429 | 1.189 | -0.7 |
| 05 | -1.11 | -1.18 | -0.084 | 0.894 |
| 06 | -1.956 | 1.05 | -0.362 | -1.19 |
| 07 | 1.167 | -2.297 | -1.143 | -1.002 |
| 08 | -0.525 | 0.271 | 0.36 | -0.41 |
| 09 | 0.113 | -0.349 | -0.096 | 0.236 |
| 10 | -0.163 | -1.348 | 1.019 | -2.121 |
| 11 | 0.749 | -2.499 | 0.523 | -2.145 |
| 12 | -1.497 | -2.868 | 1.262 | -1.683 |
| 13 | -1.644 | -1.939 | 0.601 | 0.609 |
| 14 | -0.646 | -1.039 | -0.065 | 0.782 |
| 15 | -1.92 | 0.835 | -0.073 | -0.479 |
| 16 | 0.987 | -1.156 | -0.33 | -1.258 |