Model info
| Transcription factor | USF1 | ||||||||
| Model | USF1_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 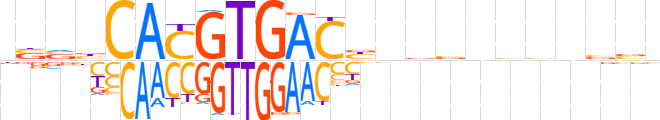 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Consensus | nvvbCACGTGACbnvvnbbnvbb | ||||||||
| wAUC | 0.9825472442714138 | ||||||||
| Best AUC | 0.9892244682038108 | ||||||||
| Benchmark datasets | 14 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | bHLH-ZIP factors{1.2.6} | ||||||||
| TF subfamily | USF factors{1.2.6.2} | ||||||||
| HGNC | 12593 | ||||||||
| EntrezGene | 7391 | ||||||||
| UniProt ID | USF1_HUMAN | ||||||||
| UniProt AC | P22415 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 6.331 | 10.64 | 47.898 | 6.126 | 24.727 | 54.095 | 61.797 | 21.516 | 11.14 | 45.842 | 79.666 | 7.64 | 7.931 | 15.439 | 89.461 | 9.753 |
| 02 | 11.125 | 9.566 | 29.437 | 0.0 | 48.936 | 32.51 | 32.287 | 12.283 | 40.366 | 67.865 | 161.663 | 8.927 | 7.283 | 11.297 | 26.454 | 0.0 |
| 03 | 7.657 | 40.157 | 15.937 | 43.959 | 6.272 | 66.705 | 14.116 | 34.145 | 20.226 | 93.983 | 53.285 | 82.347 | 0.94 | 9.676 | 3.227 | 7.367 |
| 04 | 0.0 | 35.096 | 0.0 | 0.0 | 0.0 | 209.733 | 0.788 | 0.0 | 0.0 | 86.566 | 0.0 | 0.0 | 0.0 | 166.824 | 0.995 | 0.0 |
| 05 | 0.0 | 0.0 | 0.0 | 0.0 | 493.051 | 0.0 | 4.176 | 0.991 | 0.995 | 0.0 | 0.0 | 0.788 | 0.0 | 0.0 | 0.0 | 0.0 |
| 06 | 0.0 | 391.901 | 6.859 | 95.285 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.831 | 1.311 | 1.034 | 0.0 | 1.778 | 0.0 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 19.003 | 4.341 | 372.166 | 0.0 | 0.0 | 0.808 | 7.362 | 0.0 | 3.766 | 0.0 | 92.554 | 0.0 |
| 08 | 0.831 | 0.0 | 0.899 | 21.039 | 0.0 | 0.0 | 0.0 | 5.149 | 0.0 | 0.831 | 0.727 | 470.525 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 0.0 | 0.831 | 0.0 | 0.0 | 0.0 | 0.0 | 0.831 | 0.0 | 0.0 | 0.0 | 1.626 | 0.0 | 0.0 | 0.0 | 496.712 | 0.0 |
| 10 | 0.0 | 0.0 | 0.0 | 0.0 | 0.831 | 0.0 | 0.0 | 0.0 | 466.091 | 11.822 | 19.286 | 1.971 | 0.0 | 0.0 | 0.0 | 0.0 |
| 11 | 0.0 | 423.463 | 4.873 | 38.585 | 1.128 | 7.747 | 0.0 | 2.946 | 0.0 | 10.399 | 1.568 | 7.32 | 0.0 | 0.801 | 1.17 | 0.0 |
| 12 | 0.0 | 0.0 | 1.128 | 0.0 | 35.903 | 231.828 | 62.462 | 112.217 | 0.0 | 3.753 | 3.857 | 0.0 | 0.0 | 20.711 | 15.141 | 12.999 |
| 13 | 8.643 | 15.012 | 11.158 | 1.091 | 58.46 | 101.049 | 40.43 | 56.354 | 8.65 | 44.177 | 23.465 | 6.296 | 21.682 | 37.222 | 41.99 | 24.322 |
| 14 | 21.842 | 31.831 | 32.442 | 11.32 | 42.177 | 45.968 | 77.865 | 31.449 | 13.258 | 32.181 | 46.405 | 25.199 | 9.204 | 22.457 | 45.314 | 11.09 |
| 15 | 13.199 | 15.984 | 46.737 | 10.561 | 29.603 | 42.884 | 37.406 | 22.543 | 34.486 | 66.616 | 79.727 | 21.196 | 4.78 | 26.666 | 36.76 | 10.851 |
| 16 | 15.774 | 15.281 | 39.287 | 11.727 | 28.855 | 45.392 | 51.267 | 26.636 | 24.823 | 71.528 | 66.674 | 37.605 | 13.083 | 11.806 | 15.633 | 24.629 |
| 17 | 21.177 | 19.283 | 29.07 | 13.006 | 11.455 | 57.014 | 40.629 | 34.908 | 27.207 | 69.098 | 53.627 | 22.929 | 5.019 | 40.848 | 21.256 | 33.474 |
| 18 | 13.893 | 17.027 | 28.148 | 5.79 | 22.052 | 66.026 | 35.292 | 62.872 | 14.59 | 57.605 | 51.145 | 21.243 | 5.803 | 44.237 | 32.41 | 21.866 |
| 19 | 16.584 | 12.469 | 21.941 | 5.345 | 27.54 | 71.498 | 36.828 | 49.029 | 23.029 | 51.963 | 53.354 | 18.65 | 9.603 | 30.688 | 51.676 | 19.804 |
| 20 | 17.198 | 15.586 | 39.983 | 3.987 | 21.42 | 52.928 | 58.877 | 33.393 | 20.738 | 48.959 | 76.438 | 17.664 | 9.857 | 25.281 | 46.149 | 11.54 |
| 21 | 3.878 | 32.339 | 26.916 | 6.08 | 23.333 | 55.432 | 36.905 | 27.084 | 18.41 | 110.201 | 70.57 | 22.267 | 3.734 | 28.099 | 28.366 | 6.386 |
| 22 | 6.998 | 24.795 | 13.002 | 4.561 | 31.815 | 105.661 | 44.834 | 43.761 | 20.652 | 77.384 | 47.09 | 17.631 | 8.962 | 22.396 | 12.596 | 17.862 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.549 | -1.054 | 0.423 | -1.58 | -0.231 | 0.544 | 0.676 | -0.368 | -1.01 | 0.379 | 0.928 | -1.371 | -1.336 | -0.693 | 1.044 | -1.138 |
| 02 | -1.011 | -1.156 | -0.059 | -4.4 | 0.444 | 0.039 | 0.032 | -0.915 | 0.253 | 0.769 | 1.634 | -1.223 | -1.417 | -0.996 | -0.164 | -4.4 |
| 03 | -1.369 | 0.248 | -0.662 | 0.338 | -1.558 | 0.752 | -0.78 | 0.088 | -0.428 | 1.093 | 0.529 | 0.961 | -3.17 | -1.145 | -2.169 | -1.406 |
| 04 | -4.4 | 0.115 | -4.4 | -4.4 | -4.4 | 1.893 | -3.292 | -4.4 | -4.4 | 1.011 | -4.4 | -4.4 | -4.4 | 1.665 | -3.13 | -4.4 |
| 05 | -4.4 | -4.4 | -4.4 | -4.4 | 2.747 | -4.4 | -1.936 | -3.133 | -3.13 | -4.4 | -4.4 | -3.292 | -4.4 | -4.4 | -4.4 | -4.4 |
| 06 | -4.4 | 2.518 | -1.474 | 1.107 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -2.657 | -2.924 | -3.102 | -4.4 | -2.681 | -4.4 | -4.4 |
| 07 | -4.4 | -4.4 | -4.4 | -4.4 | -0.49 | -1.901 | 2.466 | -4.4 | -4.4 | -3.275 | -1.407 | -4.4 | -2.03 | -4.4 | 1.078 | -4.4 |
| 08 | -3.256 | -4.4 | -3.202 | -0.39 | -4.4 | -4.4 | -4.4 | -1.743 | -4.4 | -3.256 | -3.345 | 2.7 | -4.4 | -4.4 | -4.4 | -4.4 |
| 09 | -4.4 | -3.256 | -4.4 | -4.4 | -4.4 | -4.4 | -3.256 | -4.4 | -4.4 | -4.4 | -2.754 | -4.4 | -4.4 | -4.4 | 2.754 | -4.4 |
| 10 | -4.4 | -4.4 | -4.4 | -4.4 | -3.256 | -4.4 | -4.4 | -4.4 | 2.691 | -0.952 | -0.475 | -2.596 | -4.4 | -4.4 | -4.4 | -4.4 |
| 11 | -4.4 | 2.595 | -1.794 | 0.209 | -3.038 | -1.358 | -4.4 | -2.25 | -4.4 | -1.076 | -2.783 | -1.412 | -4.4 | -3.281 | -3.011 | -4.4 |
| 12 | -4.4 | -4.4 | -3.038 | -4.4 | 0.137 | 1.993 | 0.686 | 1.27 | -4.4 | -2.033 | -2.009 | -4.4 | -4.4 | -0.405 | -0.712 | -0.86 |
| 13 | -1.254 | -0.72 | -1.008 | -3.063 | 0.621 | 1.165 | 0.255 | 0.584 | -1.253 | 0.343 | -0.282 | -1.555 | -0.36 | 0.173 | 0.292 | -0.247 |
| 14 | -0.353 | 0.018 | 0.037 | -0.994 | 0.297 | 0.382 | 0.906 | 0.006 | -0.841 | 0.029 | 0.391 | -0.212 | -1.193 | -0.326 | 0.368 | -1.014 |
| 15 | -0.845 | -0.659 | 0.398 | -1.061 | -0.053 | 0.313 | 0.178 | -0.322 | 0.097 | 0.75 | 0.929 | -0.382 | -1.812 | -0.157 | 0.161 | -1.035 |
| 16 | -0.672 | -0.703 | 0.226 | -0.96 | -0.079 | 0.369 | 0.49 | -0.158 | -0.227 | 0.821 | 0.751 | 0.183 | -0.854 | -0.953 | -0.68 | -0.235 |
| 17 | -0.383 | -0.475 | -0.071 | -0.86 | -0.983 | 0.596 | 0.26 | 0.109 | -0.137 | 0.787 | 0.535 | -0.305 | -1.767 | 0.265 | -0.38 | 0.068 |
| 18 | -0.795 | -0.597 | -0.103 | -1.633 | -0.343 | 0.742 | 0.12 | 0.693 | -0.748 | 0.606 | 0.488 | -0.38 | -1.631 | 0.344 | 0.036 | -0.352 |
| 19 | -0.623 | -0.9 | -0.348 | -1.708 | -0.125 | 0.821 | 0.162 | 0.446 | -0.301 | 0.504 | 0.53 | -0.508 | -1.153 | -0.018 | 0.498 | -0.449 |
| 20 | -0.587 | -0.683 | 0.244 | -1.978 | -0.372 | 0.522 | 0.628 | 0.066 | -0.404 | 0.445 | 0.887 | -0.561 | -1.128 | -0.209 | 0.386 | -0.975 |
| 21 | -2.004 | 0.034 | -0.147 | -1.587 | -0.288 | 0.568 | 0.164 | -0.141 | -0.521 | 1.251 | 0.808 | -0.334 | -2.038 | -0.105 | -0.096 | -1.541 |
| 22 | -1.455 | -0.228 | -0.86 | -1.855 | 0.018 | 1.21 | 0.357 | 0.333 | -0.408 | 0.899 | 0.406 | -0.563 | -1.219 | -0.328 | -0.891 | -0.55 |