Model info
| Transcription factor | THAP1 | ||||||||
| Model | THAP1_HUMAN.H10DI.D | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 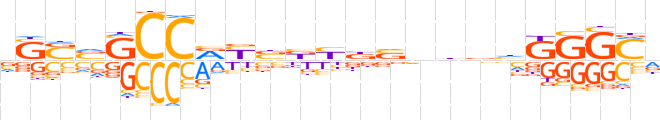 | ||||||||
| LOGO (reverse complement) | 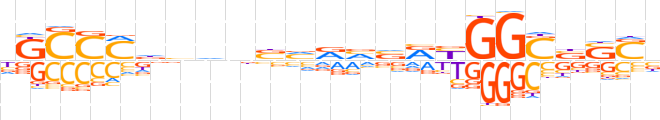 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | D | ||||||||
| Consensus | vSSvSCCRYbbYbvnbbhKGGMv | ||||||||
| wAUC | 0.6973007506838194 | ||||||||
| Best AUC | 0.6973007506838194 | ||||||||
| Benchmark datasets | 1 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | THAP-related factors{2.9.1} | ||||||||
| TF subfamily | THAP1{2.9.1.0.1} | ||||||||
| HGNC | 20856 | ||||||||
| EntrezGene | 55145 | ||||||||
| UniProt ID | THAP1_HUMAN | ||||||||
| UniProt AC | Q9NVV9 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 4.21 | 5.152 | 69.79 | 7.888 | 14.238 | 21.732 | 120.444 | 14.185 | 12.479 | 18.787 | 131.987 | 9.356 | 1.188 | 16.252 | 46.156 | 6.158 |
| 02 | 5.098 | 18.655 | 5.905 | 2.456 | 4.565 | 46.91 | 6.904 | 3.543 | 26.202 | 242.825 | 74.896 | 24.454 | 0.0 | 25.727 | 8.094 | 3.767 |
| 03 | 10.17 | 13.424 | 11.018 | 1.252 | 52.364 | 197.722 | 59.145 | 24.885 | 13.683 | 46.5 | 25.999 | 9.617 | 3.746 | 22.777 | 5.633 | 2.064 |
| 04 | 3.529 | 2.806 | 70.838 | 2.79 | 26.897 | 24.785 | 206.521 | 22.22 | 3.859 | 15.119 | 79.351 | 3.467 | 2.357 | 1.272 | 31.174 | 3.015 |
| 05 | 0.0 | 33.864 | 2.778 | 0.0 | 0.0 | 42.282 | 0.842 | 0.859 | 4.946 | 369.594 | 5.978 | 7.366 | 0.0 | 25.914 | 2.886 | 2.69 |
| 06 | 1.239 | 2.814 | 0.893 | 0.0 | 22.522 | 434.999 | 12.065 | 2.069 | 1.818 | 9.802 | 0.865 | 0.0 | 1.089 | 9.175 | 0.652 | 0.0 |
| 07 | 2.705 | 7.298 | 15.976 | 0.688 | 286.135 | 69.288 | 78.795 | 22.572 | 6.802 | 3.287 | 3.267 | 1.119 | 1.226 | 0.0 | 0.0 | 0.843 |
| 08 | 4.279 | 42.441 | 24.772 | 225.376 | 3.128 | 35.326 | 15.829 | 25.59 | 1.61 | 55.328 | 16.78 | 24.319 | 0.775 | 6.49 | 11.926 | 6.03 |
| 09 | 0.0 | 4.166 | 3.591 | 2.034 | 13.648 | 66.812 | 49.009 | 10.117 | 7.579 | 26.262 | 33.809 | 1.658 | 14.952 | 148.733 | 74.893 | 42.737 |
| 10 | 2.207 | 1.21 | 17.644 | 15.118 | 6.414 | 34.12 | 59.182 | 146.258 | 18.81 | 37.993 | 46.337 | 58.163 | 2.519 | 8.022 | 6.491 | 39.513 |
| 11 | 1.043 | 15.442 | 3.296 | 10.17 | 7.967 | 46.322 | 4.917 | 22.138 | 15.817 | 89.53 | 10.551 | 13.755 | 3.693 | 40.822 | 14.39 | 200.146 |
| 12 | 0.767 | 0.737 | 26.222 | 0.795 | 13.563 | 45.911 | 94.265 | 38.377 | 5.17 | 2.098 | 22.91 | 2.977 | 21.354 | 24.272 | 125.961 | 74.621 |
| 13 | 12.844 | 6.889 | 17.507 | 3.613 | 7.353 | 25.058 | 31.457 | 9.151 | 34.599 | 64.376 | 157.01 | 13.373 | 13.252 | 30.692 | 63.601 | 9.225 |
| 14 | 2.868 | 30.007 | 26.941 | 8.232 | 12.735 | 59.232 | 22.483 | 32.565 | 74.511 | 88.708 | 56.631 | 49.724 | 4.928 | 16.165 | 8.763 | 5.507 |
| 15 | 32.637 | 8.165 | 28.688 | 25.551 | 19.23 | 56.212 | 28.469 | 90.2 | 13.298 | 32.958 | 38.174 | 30.388 | 7.311 | 18.513 | 6.843 | 63.362 |
| 16 | 8.768 | 10.999 | 45.161 | 7.548 | 14.875 | 53.568 | 28.598 | 18.807 | 15.792 | 34.256 | 36.738 | 15.389 | 31.994 | 53.166 | 79.219 | 45.124 |
| 17 | 20.711 | 22.137 | 15.876 | 12.704 | 20.202 | 99.552 | 13.526 | 18.708 | 49.972 | 76.451 | 19.853 | 43.44 | 13.41 | 36.439 | 15.927 | 21.092 |
| 18 | 1.024 | 6.33 | 88.997 | 7.944 | 9.224 | 14.064 | 142.362 | 68.929 | 0.893 | 3.892 | 54.153 | 6.244 | 3.618 | 3.37 | 75.677 | 13.278 |
| 19 | 2.036 | 2.561 | 10.162 | 0.0 | 0.787 | 9.454 | 13.826 | 3.589 | 7.232 | 65.462 | 283.697 | 4.798 | 4.504 | 10.208 | 78.52 | 3.164 |
| 20 | 0.913 | 2.656 | 10.99 | 0.0 | 2.267 | 28.271 | 51.991 | 5.155 | 2.856 | 36.043 | 337.302 | 10.005 | 0.0 | 3.833 | 5.584 | 2.134 |
| 21 | 0.802 | 4.541 | 0.693 | 0.0 | 3.328 | 53.963 | 4.805 | 8.706 | 35.248 | 315.442 | 31.332 | 23.845 | 1.371 | 12.841 | 1.061 | 2.022 |
| 22 | 12.708 | 9.065 | 11.357 | 7.62 | 152.362 | 69.019 | 113.215 | 52.19 | 10.903 | 17.511 | 5.028 | 4.449 | 8.851 | 4.798 | 16.537 | 4.386 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.929 | -1.742 | 0.797 | -1.341 | -0.772 | -0.358 | 1.34 | -0.775 | -0.9 | -0.501 | 1.431 | -1.178 | -2.999 | -0.643 | 0.386 | -1.575 |
| 02 | -1.752 | -0.508 | -1.615 | -2.409 | -1.854 | 0.402 | -1.468 | -2.085 | -0.174 | 2.04 | 0.867 | -0.242 | -4.4 | -0.192 | -1.316 | -2.03 |
| 03 | -1.097 | -0.829 | -1.02 | -2.96 | 0.511 | 1.834 | 0.632 | -0.225 | -0.81 | 0.393 | -0.181 | -1.151 | -2.035 | -0.312 | -1.659 | -2.557 |
| 04 | -2.089 | -2.293 | 0.811 | -2.298 | -0.148 | -0.229 | 1.878 | -0.336 | -2.008 | -0.713 | 0.924 | -2.105 | -2.444 | -2.947 | -0.002 | -2.23 |
| 05 | -4.4 | 0.079 | -2.302 | -4.4 | -4.4 | 0.299 | -3.247 | -3.233 | -1.78 | 2.459 | -1.603 | -1.406 | -4.4 | -0.185 | -2.268 | -2.33 |
| 06 | -2.967 | -2.29 | -3.207 | -4.4 | -0.323 | 2.622 | -0.932 | -2.555 | -2.663 | -1.133 | -3.229 | -4.4 | -3.064 | -1.196 | -3.415 | -4.4 |
| 07 | -2.325 | -1.415 | -0.659 | -3.381 | 2.203 | 0.789 | 0.917 | -0.321 | -1.482 | -2.153 | -2.158 | -3.044 | -2.975 | -4.4 | -4.4 | -3.246 |
| 08 | -1.914 | 0.303 | -0.229 | 1.965 | -2.197 | 0.121 | -0.668 | -0.197 | -2.762 | 0.566 | -0.611 | -0.247 | -3.303 | -1.526 | -0.944 | -1.595 |
| 09 | -4.4 | -1.938 | -2.073 | -2.569 | -0.813 | 0.753 | 0.446 | -1.103 | -1.379 | -0.172 | 0.078 | -2.738 | -0.724 | 1.55 | 0.867 | 0.31 |
| 10 | -2.5 | -2.985 | -0.562 | -0.713 | -1.537 | 0.087 | 0.633 | 1.534 | -0.5 | 0.193 | 0.39 | 0.616 | -2.387 | -1.325 | -1.526 | 0.232 |
| 11 | -3.095 | -0.692 | -2.15 | -1.097 | -1.331 | 0.39 | -1.786 | -0.34 | -0.669 | 1.045 | -1.062 | -0.805 | -2.048 | 0.264 | -0.761 | 1.847 |
| 12 | -3.31 | -3.336 | -0.173 | -3.286 | -0.819 | 0.381 | 1.096 | 0.203 | -1.739 | -2.543 | -0.306 | -2.241 | -0.375 | -0.249 | 1.385 | 0.863 |
| 13 | -0.872 | -1.47 | -0.57 | -2.068 | -1.408 | -0.218 | 0.007 | -1.199 | 0.101 | 0.716 | 1.604 | -0.832 | -0.841 | -0.018 | 0.704 | -1.191 |
| 14 | -2.274 | -0.04 | -0.146 | -1.3 | -0.88 | 0.634 | -0.324 | 0.041 | 0.862 | 1.035 | 0.589 | 0.46 | -1.784 | -0.648 | -1.24 | -1.68 |
| 15 | 0.043 | -1.308 | -0.084 | -0.199 | -0.478 | 0.582 | -0.092 | 1.052 | -0.838 | 0.053 | 0.198 | -0.028 | -1.413 | -0.515 | -1.476 | 0.701 |
| 16 | -1.24 | -1.022 | 0.364 | -1.383 | -0.729 | 0.534 | -0.088 | -0.5 | -0.671 | 0.091 | 0.16 | -0.696 | 0.023 | 0.526 | 0.923 | 0.364 |
| 17 | -0.405 | -0.34 | -0.665 | -0.882 | -0.43 | 1.15 | -0.821 | -0.505 | 0.465 | 0.887 | -0.447 | 0.326 | -0.83 | 0.152 | -0.662 | -0.387 |
| 18 | -3.109 | -1.55 | 1.039 | -1.334 | -1.191 | -0.784 | 1.507 | 0.784 | -3.207 | -2.0 | 0.545 | -1.562 | -2.066 | -2.13 | 0.877 | -0.839 |
| 19 | -2.569 | -2.373 | -1.098 | -4.4 | -3.292 | -1.168 | -0.8 | -2.074 | -1.424 | 0.733 | 2.195 | -1.808 | -1.867 | -1.094 | 0.914 | -2.187 |
| 20 | -3.191 | -2.341 | -1.023 | -4.4 | -2.478 | -0.099 | 0.504 | -1.742 | -2.277 | 0.141 | 2.368 | -1.113 | -4.4 | -2.014 | -1.667 | -2.529 |
| 21 | -3.28 | -1.859 | -3.376 | -4.4 | -2.142 | 0.541 | -1.807 | -1.247 | 0.119 | 2.301 | 0.003 | -0.267 | -2.889 | -0.872 | -3.083 | -2.575 |
| 22 | -0.882 | -1.208 | -0.991 | -1.374 | 1.574 | 0.786 | 1.278 | 0.508 | -1.03 | -0.57 | -1.765 | -1.878 | -1.231 | -1.808 | -0.626 | -1.891 |