Model info
| Transcription factor | TCF7L2 | ||||||||
| Model | TF7L2_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 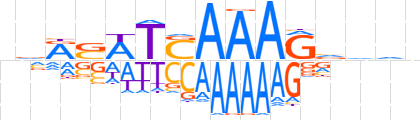 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 15 | ||||||||
| Quality | A | ||||||||
| Consensus | ndbbCTTTSAWSYbn | ||||||||
| wAUC | 0.7927364419761587 | ||||||||
| Best AUC | 0.8641073080716737 | ||||||||
| Benchmark datasets | 12 | ||||||||
| Aligned words | 497 | ||||||||
| TF family | TCF-7-related factors{4.1.3} | ||||||||
| TF subfamily | TCF-7L2 (TCF-4) [1]{4.1.3.0.3} | ||||||||
| HGNC | 11641 | ||||||||
| EntrezGene | 6934 | ||||||||
| UniProt ID | TF7L2_HUMAN | ||||||||
| UniProt AC | Q9NQB0 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 33.518 | 14.244 | 27.919 | 54.487 | 27.692 | 10.684 | 6.467 | 43.58 | 16.997 | 37.821 | 38.286 | 44.734 | 13.068 | 24.887 | 25.229 | 74.403 |
| 02 | 16.02 | 17.404 | 29.154 | 28.697 | 13.368 | 26.654 | 7.084 | 40.53 | 11.556 | 39.293 | 22.422 | 24.63 | 16.91 | 47.656 | 57.295 | 95.344 |
| 03 | 3.666 | 26.175 | 16.422 | 11.591 | 17.855 | 46.411 | 10.969 | 55.772 | 6.152 | 55.638 | 32.725 | 21.439 | 10.009 | 72.752 | 40.11 | 66.329 |
| 04 | 1.841 | 33.378 | 0.0 | 2.462 | 9.691 | 157.518 | 2.999 | 30.767 | 1.881 | 91.549 | 0.902 | 5.895 | 4.056 | 111.178 | 17.946 | 21.952 |
| 05 | 0.0 | 0.0 | 0.0 | 17.469 | 0.884 | 2.007 | 0.0 | 390.733 | 0.945 | 0.0 | 0.0 | 20.902 | 0.0 | 0.0 | 0.0 | 61.076 |
| 06 | 0.0 | 0.884 | 0.0 | 0.945 | 0.0 | 0.0 | 0.0 | 2.007 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 18.3 | 5.727 | 466.152 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 1.38 | 0.0 | 0.0 | 17.805 | 0.0 | 0.0 | 0.828 | 4.899 | 1.745 | 0.0 | 1.827 | 465.532 |
| 08 | 0.0 | 0.0 | 2.195 | 0.93 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.656 | 0.0 | 14.688 | 84.783 | 358.942 | 29.822 |
| 09 | 12.813 | 0.0 | 0.0 | 1.875 | 77.617 | 0.0 | 0.0 | 7.166 | 311.697 | 1.377 | 8.974 | 41.745 | 26.527 | 0.802 | 0.0 | 3.423 |
| 10 | 153.142 | 2.666 | 10.77 | 262.076 | 0.0 | 0.0 | 0.0 | 2.178 | 1.828 | 0.0 | 0.0 | 7.146 | 3.861 | 1.004 | 2.357 | 46.987 |
| 11 | 8.772 | 29.057 | 118.382 | 2.62 | 0.948 | 2.722 | 0.0 | 0.0 | 0.854 | 1.84 | 8.211 | 2.223 | 7.385 | 193.215 | 101.43 | 16.357 |
| 12 | 3.81 | 0.0 | 3.408 | 10.741 | 21.833 | 37.033 | 1.351 | 166.617 | 25.594 | 24.247 | 22.351 | 155.831 | 0.826 | 2.813 | 0.0 | 17.561 |
| 13 | 4.768 | 20.523 | 10.342 | 16.429 | 12.311 | 23.941 | 5.716 | 22.125 | 4.292 | 6.815 | 8.862 | 7.141 | 28.102 | 95.973 | 97.911 | 128.765 |
| 14 | 13.449 | 17.779 | 13.337 | 4.907 | 50.282 | 23.337 | 7.749 | 65.883 | 24.883 | 35.96 | 33.291 | 28.697 | 20.689 | 38.573 | 40.678 | 74.519 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.081 | -0.759 | -0.099 | 0.563 | -0.107 | -1.038 | -1.518 | 0.341 | -0.587 | 0.201 | 0.213 | 0.367 | -0.843 | -0.213 | -0.199 | 0.872 |
| 02 | -0.645 | -0.564 | -0.057 | -0.072 | -0.821 | -0.145 | -1.431 | 0.269 | -0.962 | 0.238 | -0.315 | -0.223 | -0.592 | 0.43 | 0.613 | 1.119 |
| 03 | -2.043 | -0.163 | -0.621 | -0.959 | -0.539 | 0.403 | -1.013 | 0.586 | -1.565 | 0.583 | 0.057 | -0.359 | -1.101 | 0.85 | 0.259 | 0.758 |
| 04 | -2.641 | 0.077 | -4.39 | -2.395 | -1.132 | 1.62 | -2.223 | -0.003 | -2.623 | 1.079 | -3.188 | -1.605 | -1.951 | 1.272 | -0.534 | -0.336 |
| 05 | -4.39 | -4.39 | -4.39 | -0.56 | -3.202 | -2.569 | -4.39 | 2.527 | -3.155 | -4.39 | -4.39 | -0.384 | -4.39 | -4.39 | -4.39 | 0.676 |
| 06 | -4.39 | -3.202 | -4.39 | -3.155 | -4.39 | -4.39 | -4.39 | -2.569 | -4.39 | -4.39 | -4.39 | -4.39 | -4.39 | -0.515 | -1.632 | 2.703 |
| 07 | -4.39 | -4.39 | -4.39 | -4.39 | -2.873 | -4.39 | -4.39 | -0.541 | -4.39 | -4.39 | -3.247 | -1.777 | -2.685 | -4.39 | -2.647 | 2.702 |
| 08 | -4.39 | -4.39 | -2.494 | -3.167 | -4.39 | -4.39 | -4.39 | -4.39 | -4.39 | -4.39 | -2.329 | -4.39 | -0.729 | 1.002 | 2.442 | -0.034 |
| 09 | -0.862 | -4.39 | -4.39 | -2.626 | 0.914 | -4.39 | -4.39 | -1.42 | 2.301 | -2.875 | -1.206 | 0.298 | -0.15 | -3.269 | -4.39 | -2.105 |
| 10 | 1.591 | -2.326 | -1.03 | 2.128 | -4.39 | -4.39 | -4.39 | -2.5 | -2.647 | -4.39 | -4.39 | -1.423 | -1.996 | -3.112 | -2.433 | 0.416 |
| 11 | -1.228 | -0.06 | 1.335 | -2.341 | -3.153 | -2.308 | -4.39 | -4.39 | -3.226 | -2.641 | -1.291 | -2.483 | -1.392 | 1.823 | 1.181 | -0.624 |
| 12 | -2.008 | -4.39 | -2.109 | -1.033 | -0.341 | 0.18 | -2.889 | 1.676 | -0.185 | -0.238 | -0.318 | 1.609 | -3.249 | -2.279 | -4.39 | -0.555 |
| 13 | -1.802 | -0.402 | -1.069 | -0.62 | -0.901 | -0.251 | -1.634 | -0.328 | -1.899 | -1.468 | -1.218 | -1.424 | -0.093 | 1.126 | 1.146 | 1.419 |
| 14 | -0.815 | -0.543 | -0.823 | -1.776 | 0.483 | -0.276 | -1.346 | 0.751 | -0.213 | 0.151 | 0.074 | -0.072 | -0.394 | 0.22 | 0.273 | 0.874 |