Model info
| Transcription factor | TEAD4 | ||||||||
| Model | TEAD4_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 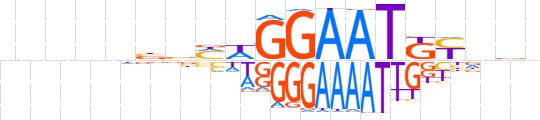 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 19 | ||||||||
| Quality | A | ||||||||
| Consensus | nhnRMATTCCWKvvnnnnn | ||||||||
| wAUC | 0.7982976703816592 | ||||||||
| Best AUC | 0.8851818058295589 | ||||||||
| Benchmark datasets | 9 | ||||||||
| Aligned words | 503 | ||||||||
| TF family | TEF-1-related factors{3.6.1} | ||||||||
| TF subfamily | TEF-3 (TEAD-4, TCF-13L1){3.6.1.0.2} | ||||||||
| HGNC | 11717 | ||||||||
| EntrezGene | 7004 | ||||||||
| UniProt ID | TEAD4_HUMAN | ||||||||
| UniProt AC | Q15561 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 23.52 | 66.424 | 19.188 | 13.732 | 26.191 | 54.706 | 5.239 | 39.258 | 22.677 | 34.26 | 31.663 | 28.142 | 16.597 | 45.128 | 19.005 | 48.886 |
| 02 | 32.552 | 19.348 | 20.413 | 16.672 | 23.625 | 74.6 | 13.976 | 88.318 | 16.117 | 31.496 | 13.473 | 14.009 | 17.203 | 41.296 | 46.112 | 25.407 |
| 03 | 48.017 | 0.0 | 34.723 | 6.758 | 120.3 | 0.954 | 40.59 | 4.895 | 41.715 | 0.0 | 46.379 | 5.88 | 31.685 | 0.893 | 105.794 | 6.034 |
| 04 | 79.915 | 142.264 | 18.316 | 1.223 | 0.0 | 1.847 | 0.0 | 0.0 | 44.098 | 171.099 | 8.26 | 4.029 | 5.513 | 17.088 | 0.966 | 0.0 |
| 05 | 125.706 | 3.032 | 0.788 | 0.0 | 328.735 | 3.563 | 0.0 | 0.0 | 27.542 | 0.0 | 0.0 | 0.0 | 3.363 | 0.0 | 0.854 | 1.036 |
| 06 | 0.0 | 12.773 | 8.445 | 464.127 | 0.0 | 0.0 | 0.0 | 6.594 | 0.0 | 0.0 | 0.0 | 1.642 | 0.0 | 0.0 | 0.0 | 1.036 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 12.773 | 0.0 | 0.0 | 0.0 | 8.445 | 21.533 | 0.0 | 2.028 | 449.839 |
| 08 | 0.0 | 20.542 | 0.99 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.028 | 0.0 | 0.0 | 2.075 | 447.447 | 1.76 | 19.775 |
| 09 | 0.0 | 2.075 | 0.0 | 0.0 | 2.061 | 418.38 | 0.0 | 49.576 | 0.0 | 2.751 | 0.0 | 0.0 | 0.0 | 14.353 | 0.0 | 5.422 |
| 10 | 0.0 | 1.056 | 0.0 | 1.005 | 216.386 | 18.329 | 20.371 | 182.473 | 0.0 | 0.0 | 0.0 | 0.0 | 2.958 | 3.929 | 4.731 | 43.38 |
| 11 | 20.151 | 19.835 | 158.502 | 20.857 | 10.817 | 3.989 | 6.207 | 2.301 | 1.805 | 7.087 | 15.287 | 0.922 | 26.483 | 26.945 | 126.654 | 46.776 |
| 12 | 9.009 | 16.869 | 26.092 | 7.287 | 22.274 | 12.111 | 11.15 | 12.32 | 54.964 | 114.249 | 114.082 | 23.355 | 9.382 | 24.389 | 18.662 | 18.423 |
| 13 | 21.451 | 27.747 | 36.858 | 9.573 | 25.067 | 106.629 | 14.746 | 21.176 | 41.081 | 61.667 | 59.457 | 7.781 | 9.967 | 25.675 | 18.323 | 7.42 |
| 14 | 37.513 | 19.115 | 30.131 | 10.807 | 101.383 | 33.429 | 7.355 | 79.551 | 42.507 | 28.493 | 47.746 | 10.639 | 5.423 | 11.32 | 18.215 | 10.991 |
| 15 | 20.003 | 34.685 | 29.513 | 102.626 | 23.975 | 23.364 | 7.714 | 37.303 | 20.36 | 31.187 | 38.427 | 13.474 | 10.952 | 22.758 | 59.177 | 19.103 |
| 16 | 12.906 | 28.869 | 23.243 | 10.271 | 23.194 | 32.897 | 8.698 | 47.204 | 17.818 | 31.923 | 61.27 | 23.819 | 26.863 | 31.425 | 41.403 | 72.815 |
| 17 | 17.156 | 20.761 | 18.447 | 24.417 | 35.642 | 40.82 | 11.227 | 37.426 | 17.568 | 35.857 | 44.165 | 37.025 | 20.449 | 66.287 | 28.165 | 39.208 |
| 18 | 20.132 | 20.787 | 44.649 | 5.248 | 33.461 | 74.474 | 8.879 | 46.91 | 14.954 | 31.649 | 25.992 | 29.409 | 16.772 | 31.5 | 48.815 | 40.989 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.269 | 0.758 | -0.469 | -0.796 | -0.164 | 0.565 | -1.716 | 0.236 | -0.305 | 0.102 | 0.024 | -0.093 | -0.611 | 0.374 | -0.479 | 0.454 |
| 02 | 0.051 | -0.461 | -0.409 | -0.607 | -0.265 | 0.874 | -0.779 | 1.042 | -0.64 | 0.018 | -0.815 | -0.777 | -0.576 | 0.286 | 0.396 | -0.193 |
| 03 | 0.436 | -4.391 | 0.115 | -1.477 | 1.35 | -3.15 | 0.269 | -1.779 | 0.296 | -4.391 | 0.402 | -1.608 | 0.024 | -3.196 | 1.221 | -1.584 |
| 04 | 0.942 | 1.517 | -0.515 | -2.967 | -4.391 | -2.639 | -4.391 | -4.391 | 0.352 | 1.701 | -1.286 | -1.958 | -1.669 | -0.583 | -3.141 | -4.391 |
| 05 | 1.393 | -2.214 | -3.282 | -4.391 | 2.353 | -2.07 | -4.391 | -4.391 | -0.114 | -4.391 | -4.391 | -4.391 | -2.122 | -4.391 | -3.227 | -3.091 |
| 06 | -4.391 | -0.866 | -1.265 | 2.697 | -4.391 | -4.391 | -4.391 | -1.5 | -4.391 | -4.391 | -4.391 | -2.736 | -4.391 | -4.391 | -4.391 | -3.091 |
| 07 | -4.391 | -4.391 | -4.391 | -4.391 | -4.391 | -4.391 | -4.391 | -0.866 | -4.391 | -4.391 | -4.391 | -1.265 | -0.356 | -4.391 | -2.562 | 2.666 |
| 08 | -4.391 | -0.402 | -3.123 | -4.391 | -4.391 | -4.391 | -4.391 | -4.391 | -4.391 | -2.562 | -4.391 | -4.391 | -2.542 | 2.661 | -2.679 | -0.44 |
| 09 | -4.391 | -2.542 | -4.391 | -4.391 | -2.548 | 2.594 | -4.391 | 0.468 | -4.391 | -2.3 | -4.391 | -4.391 | -4.391 | -0.753 | -4.391 | -1.684 |
| 10 | -4.391 | -3.076 | -4.391 | -3.112 | 1.935 | -0.514 | -0.411 | 1.765 | -4.391 | -4.391 | -4.391 | -4.391 | -2.236 | -1.981 | -1.811 | 0.335 |
| 11 | -0.421 | -0.437 | 1.625 | -0.388 | -1.027 | -1.967 | -1.557 | -2.455 | -2.658 | -1.432 | -0.692 | -3.174 | -0.153 | -0.136 | 1.401 | 0.41 |
| 12 | -1.203 | -0.595 | -0.167 | -1.406 | -0.323 | -0.918 | -0.998 | -0.901 | 0.57 | 1.298 | 1.297 | -0.276 | -1.164 | -0.234 | -0.497 | -0.509 |
| 13 | -0.36 | -0.107 | 0.174 | -1.145 | -0.207 | 1.229 | -0.727 | -0.373 | 0.281 | 0.684 | 0.648 | -1.343 | -1.106 | -0.183 | -0.515 | -1.389 |
| 14 | 0.191 | -0.473 | -0.025 | -1.028 | 1.179 | 0.077 | -1.397 | 0.938 | 0.315 | -0.081 | 0.43 | -1.043 | -1.684 | -0.983 | -0.52 | -1.012 |
| 15 | -0.429 | 0.114 | -0.046 | 1.191 | -0.251 | -0.276 | -1.352 | 0.186 | -0.411 | 0.009 | 0.215 | -0.815 | -1.015 | -0.302 | 0.643 | -0.474 |
| 16 | -0.856 | -0.068 | -0.281 | -1.077 | -0.283 | 0.061 | -1.237 | 0.419 | -0.542 | 0.032 | 0.678 | -0.257 | -0.139 | 0.016 | 0.289 | 0.85 |
| 17 | -0.579 | -0.392 | -0.508 | -0.233 | 0.141 | 0.275 | -0.991 | 0.189 | -0.556 | 0.147 | 0.353 | 0.178 | -0.407 | 0.756 | -0.092 | 0.235 |
| 18 | -0.422 | -0.391 | 0.364 | -1.715 | 0.078 | 0.872 | -1.217 | 0.413 | -0.713 | 0.023 | -0.171 | -0.049 | -0.601 | 0.019 | 0.452 | 0.279 |