Model info
| Transcription factor | Tal1 | ||||||||
| Model | TAL1_MOUSE.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 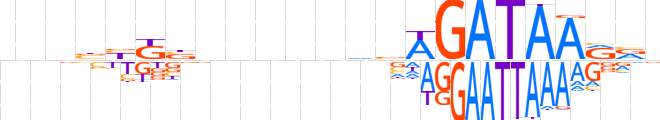 | ||||||||
| LOGO (reverse complement) | 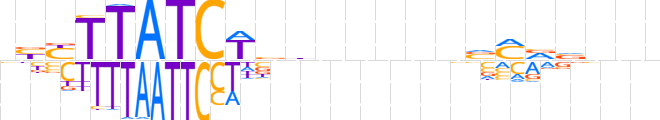 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Consensus | nnnbbKbnnnnndvWGATAARvn | ||||||||
| wAUC | 0.920395509856274 | ||||||||
| Best AUC | 0.973788791908167 | ||||||||
| Benchmark datasets | 11 | ||||||||
| Aligned words | 502 | ||||||||
| TF family | Tal-related factors{1.2.3} | ||||||||
| TF subfamily | Tal / HEN-like factors{1.2.3.1} | ||||||||
| MGI | 98480 | ||||||||
| EntrezGene | 21349 | ||||||||
| UniProt ID | TAL1_MOUSE | ||||||||
| UniProt AC | P22091 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 38.641 | 22.046 | 60.442 | 15.816 | 95.561 | 22.8 | 13.99 | 19.834 | 26.826 | 25.885 | 35.174 | 31.999 | 6.97 | 17.452 | 43.437 | 22.091 |
| 02 | 17.649 | 24.906 | 95.113 | 30.33 | 44.334 | 21.975 | 5.201 | 16.674 | 15.605 | 64.818 | 41.971 | 30.649 | 11.128 | 25.675 | 29.528 | 23.408 |
| 03 | 15.849 | 24.893 | 32.344 | 15.63 | 23.359 | 73.112 | 7.944 | 32.959 | 22.789 | 97.568 | 28.681 | 22.776 | 6.272 | 59.039 | 31.656 | 4.093 |
| 04 | 17.635 | 11.875 | 16.266 | 22.494 | 14.681 | 39.804 | 4.902 | 195.224 | 4.026 | 35.907 | 12.302 | 48.391 | 4.744 | 34.954 | 27.872 | 7.888 |
| 05 | 7.571 | 3.208 | 18.605 | 11.703 | 3.116 | 7.048 | 8.283 | 104.093 | 3.722 | 5.379 | 17.294 | 34.947 | 0.934 | 0.0 | 251.241 | 21.822 |
| 06 | 0.0 | 1.867 | 11.435 | 2.041 | 1.063 | 3.823 | 2.15 | 8.599 | 28.007 | 97.189 | 100.784 | 69.444 | 3.023 | 10.285 | 152.237 | 7.019 |
| 07 | 5.306 | 9.407 | 15.321 | 2.059 | 46.77 | 21.06 | 9.689 | 35.645 | 44.08 | 58.434 | 91.173 | 72.919 | 14.341 | 17.722 | 34.538 | 20.501 |
| 08 | 22.27 | 24.298 | 46.506 | 17.423 | 39.798 | 14.656 | 8.131 | 44.037 | 31.731 | 38.983 | 46.67 | 33.337 | 17.119 | 32.297 | 43.181 | 38.528 |
| 09 | 32.02 | 18.629 | 54.093 | 6.176 | 38.365 | 23.921 | 9.166 | 38.782 | 34.338 | 38.407 | 48.797 | 22.946 | 25.293 | 29.057 | 51.369 | 27.605 |
| 10 | 36.12 | 28.177 | 50.632 | 15.087 | 30.995 | 26.83 | 8.187 | 44.003 | 42.122 | 32.335 | 56.501 | 32.467 | 14.937 | 17.619 | 35.593 | 27.362 |
| 11 | 29.046 | 30.357 | 49.734 | 15.036 | 25.384 | 35.77 | 7.168 | 36.638 | 35.716 | 39.167 | 45.551 | 30.479 | 22.931 | 22.786 | 40.86 | 32.342 |
| 12 | 36.041 | 12.889 | 53.181 | 10.966 | 74.997 | 18.384 | 5.141 | 29.558 | 57.253 | 24.564 | 34.776 | 26.72 | 27.345 | 17.535 | 41.956 | 27.66 |
| 13 | 30.281 | 59.183 | 83.741 | 22.431 | 21.861 | 32.154 | 5.265 | 14.091 | 27.387 | 60.343 | 36.409 | 10.915 | 18.197 | 29.47 | 39.457 | 7.779 |
| 14 | 75.743 | 3.502 | 0.954 | 17.527 | 79.393 | 1.997 | 0.0 | 99.76 | 133.207 | 2.871 | 0.0 | 28.795 | 40.771 | 0.997 | 0.0 | 13.448 |
| 15 | 0.0 | 0.0 | 329.114 | 0.0 | 0.0 | 0.0 | 9.367 | 0.0 | 0.0 | 0.0 | 0.954 | 0.0 | 0.0 | 0.0 | 159.53 | 0.0 |
| 16 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 498.965 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 17 | 0.0 | 0.0 | 0.0 | 498.965 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 18 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 484.382 | 0.0 | 0.0 | 14.583 |
| 19 | 445.28 | 5.2 | 27.205 | 6.697 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 12.571 | 0.0 | 0.974 | 1.039 |
| 20 | 73.459 | 62.722 | 303.685 | 17.986 | 0.0 | 2.176 | 0.0 | 3.024 | 3.218 | 3.958 | 13.259 | 7.744 | 0.0 | 0.907 | 6.828 | 0.0 |
| 21 | 31.096 | 13.56 | 28.985 | 3.036 | 46.338 | 11.161 | 10.111 | 2.153 | 118.868 | 60.788 | 141.321 | 2.795 | 6.884 | 4.922 | 14.757 | 2.191 |
| 22 | 54.416 | 35.587 | 86.802 | 26.382 | 31.739 | 28.102 | 4.025 | 26.564 | 63.605 | 43.934 | 59.418 | 28.218 | 2.106 | 2.085 | 1.874 | 4.11 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.212 | -0.342 | 0.656 | -0.667 | 1.111 | -0.309 | -0.787 | -0.446 | -0.149 | -0.184 | 0.119 | 0.025 | -1.457 | -0.571 | 0.328 | -0.34 |
| 02 | -0.56 | -0.222 | 1.107 | -0.027 | 0.348 | -0.345 | -1.732 | -0.615 | -0.68 | 0.725 | 0.294 | -0.017 | -1.009 | -0.192 | -0.054 | -0.283 |
| 03 | -0.665 | -0.222 | 0.036 | -0.679 | -0.285 | 0.845 | -1.332 | 0.055 | -0.309 | 1.132 | -0.083 | -0.31 | -1.556 | 0.632 | 0.015 | -1.952 |
| 04 | -0.561 | -0.946 | -0.64 | -0.322 | -0.74 | 0.241 | -1.787 | 1.824 | -1.967 | 0.139 | -0.911 | 0.435 | -1.817 | 0.113 | -0.111 | -1.339 |
| 05 | -1.378 | -2.173 | -0.508 | -0.96 | -2.198 | -1.446 | -1.292 | 1.197 | -2.039 | -1.7 | -0.58 | 0.113 | -3.173 | -4.398 | 2.076 | -0.352 |
| 06 | -4.398 | -2.639 | -0.982 | -2.565 | -3.08 | -2.014 | -2.521 | -1.257 | -0.106 | 1.128 | 1.165 | 0.794 | -2.225 | -1.085 | 1.576 | -1.45 |
| 07 | -1.713 | -1.17 | -0.698 | -2.557 | 0.401 | -0.387 | -1.142 | 0.132 | 0.342 | 0.622 | 1.065 | 0.842 | -0.762 | -0.556 | 0.101 | -0.413 |
| 08 | -0.332 | -0.246 | 0.396 | -0.572 | 0.241 | -0.741 | -1.31 | 0.341 | 0.017 | 0.221 | 0.399 | 0.066 | -0.59 | 0.035 | 0.322 | 0.209 |
| 09 | 0.026 | -0.507 | 0.546 | -1.571 | 0.205 | -0.261 | -1.195 | 0.216 | 0.095 | 0.206 | 0.443 | -0.302 | -0.207 | -0.07 | 0.494 | -0.12 |
| 10 | 0.145 | -0.1 | 0.48 | -0.713 | -0.006 | -0.148 | -1.303 | 0.341 | 0.297 | 0.036 | 0.589 | 0.04 | -0.723 | -0.562 | 0.131 | -0.129 |
| 11 | -0.07 | -0.027 | 0.462 | -0.716 | -0.203 | 0.136 | -1.43 | 0.159 | 0.134 | 0.225 | 0.375 | -0.023 | -0.303 | -0.309 | 0.267 | 0.036 |
| 12 | 0.143 | -0.866 | 0.529 | -1.023 | 0.87 | -0.52 | -1.742 | -0.053 | 0.602 | -0.235 | 0.108 | -0.152 | -0.13 | -0.566 | 0.294 | -0.118 |
| 13 | -0.029 | 0.635 | 0.98 | -0.325 | -0.35 | 0.03 | -1.72 | -0.78 | -0.128 | 0.654 | 0.153 | -1.027 | -0.53 | -0.056 | 0.233 | -1.352 |
| 14 | 0.88 | -2.094 | -3.158 | -0.567 | 0.927 | -2.583 | -4.398 | 1.154 | 1.442 | -2.271 | -4.398 | -0.079 | 0.265 | -3.127 | -4.398 | -0.825 |
| 15 | -4.398 | -4.398 | 2.345 | -4.398 | -4.398 | -4.398 | -1.175 | -4.398 | -4.398 | -4.398 | -3.158 | -4.398 | -4.398 | -4.398 | 1.622 | -4.398 |
| 16 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | 2.761 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 |
| 17 | -4.398 | -4.398 | -4.398 | 2.761 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 |
| 18 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | 2.731 | -4.398 | -4.398 | -0.746 |
| 19 | 2.647 | -1.732 | -0.135 | -1.494 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -0.891 | -4.398 | -3.143 | -3.097 |
| 20 | 0.85 | 0.693 | 2.265 | -0.541 | -4.398 | -2.511 | -4.398 | -2.225 | -2.17 | -1.983 | -0.839 | -1.356 | -4.398 | -3.194 | -1.476 | -4.398 |
| 21 | -0.003 | -0.817 | -0.072 | -2.221 | 0.392 | -1.006 | -1.101 | -2.52 | 1.329 | 0.661 | 1.501 | -2.295 | -1.468 | -1.783 | -0.735 | -2.505 |
| 22 | 0.551 | 0.131 | 1.016 | -0.165 | 0.017 | -0.103 | -1.968 | -0.158 | 0.706 | 0.339 | 0.639 | -0.099 | -2.538 | -2.547 | -2.636 | -1.949 |