Model info
| Transcription factor | TAL1 | ||||||||
| Model | TAL1_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 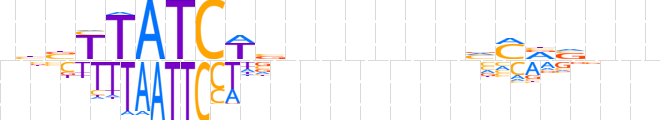 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Consensus | nnnbbKbnnnnnnbWGATAAvvn | ||||||||
| wAUC | 0.9055035222510769 | ||||||||
| Best AUC | 0.9621297718123053 | ||||||||
| Benchmark datasets | 6 | ||||||||
| Aligned words | 501 | ||||||||
| TF family | Tal-related factors{1.2.3} | ||||||||
| TF subfamily | Tal / HEN-like factors{1.2.3.1} | ||||||||
| HGNC | 11556 | ||||||||
| EntrezGene | 6886 | ||||||||
| UniProt ID | TAL1_HUMAN | ||||||||
| UniProt AC | P17542 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 29.426 | 22.339 | 57.183 | 10.961 | 69.719 | 39.979 | 13.204 | 29.945 | 25.708 | 37.148 | 38.731 | 22.566 | 6.155 | 20.503 | 58.637 | 16.772 |
| 02 | 16.577 | 18.477 | 62.987 | 32.967 | 54.505 | 24.054 | 9.35 | 32.061 | 26.815 | 68.999 | 46.716 | 25.226 | 7.659 | 21.639 | 28.606 | 22.34 |
| 03 | 8.992 | 41.044 | 38.7 | 16.819 | 18.393 | 74.819 | 6.706 | 33.251 | 5.748 | 89.052 | 31.405 | 21.455 | 5.556 | 69.175 | 28.938 | 8.924 |
| 04 | 0.941 | 7.459 | 11.411 | 18.878 | 31.818 | 34.017 | 3.923 | 204.332 | 6.024 | 33.524 | 8.778 | 57.422 | 5.29 | 22.866 | 41.13 | 11.162 |
| 05 | 0.0 | 5.887 | 29.237 | 8.949 | 3.086 | 3.462 | 6.3 | 85.019 | 5.303 | 6.917 | 21.864 | 31.159 | 5.386 | 4.346 | 269.819 | 12.242 |
| 06 | 2.342 | 1.853 | 7.015 | 2.565 | 6.329 | 4.666 | 0.93 | 8.687 | 31.953 | 85.809 | 118.363 | 91.094 | 1.284 | 6.151 | 115.782 | 14.153 |
| 07 | 10.039 | 7.666 | 22.954 | 1.25 | 28.819 | 22.479 | 4.546 | 42.634 | 28.426 | 73.87 | 73.56 | 66.234 | 16.467 | 21.889 | 45.909 | 32.235 |
| 08 | 18.597 | 20.117 | 37.105 | 7.932 | 52.011 | 18.503 | 12.01 | 43.38 | 36.83 | 54.822 | 35.546 | 19.771 | 17.071 | 30.467 | 59.63 | 35.185 |
| 09 | 33.021 | 20.742 | 62.143 | 8.602 | 38.836 | 36.973 | 10.237 | 37.862 | 23.084 | 37.632 | 60.255 | 23.32 | 8.436 | 31.986 | 35.511 | 30.335 |
| 10 | 16.435 | 17.932 | 54.732 | 14.278 | 28.996 | 36.842 | 6.122 | 55.374 | 32.424 | 49.382 | 57.577 | 28.763 | 7.793 | 24.877 | 43.241 | 24.207 |
| 11 | 13.99 | 24.205 | 36.474 | 10.98 | 39.092 | 48.352 | 7.909 | 33.68 | 37.995 | 42.909 | 52.517 | 28.252 | 12.37 | 38.968 | 38.205 | 33.081 |
| 12 | 29.587 | 13.929 | 52.881 | 7.048 | 60.443 | 27.934 | 9.881 | 56.177 | 38.592 | 18.022 | 57.451 | 21.039 | 21.476 | 14.069 | 53.343 | 17.103 |
| 13 | 27.821 | 65.061 | 44.67 | 12.546 | 7.413 | 33.479 | 12.081 | 20.981 | 16.177 | 86.37 | 55.36 | 15.648 | 6.738 | 34.469 | 41.628 | 18.532 |
| 14 | 39.279 | 2.796 | 0.0 | 16.075 | 122.564 | 7.134 | 0.835 | 88.845 | 104.878 | 4.554 | 4.219 | 40.088 | 51.084 | 0.0 | 1.258 | 15.366 |
| 15 | 0.0 | 0.0 | 317.805 | 0.0 | 0.0 | 0.0 | 14.485 | 0.0 | 0.0 | 0.0 | 6.312 | 0.0 | 0.0 | 0.0 | 160.374 | 0.0 |
| 16 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 498.085 | 0.0 | 0.891 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 17 | 0.0 | 0.822 | 1.069 | 496.194 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.891 | 0.0 | 0.0 | 0.0 | 0.0 |
| 18 | 0.0 | 0.0 | 0.0 | 0.0 | 0.822 | 0.0 | 0.0 | 0.0 | 1.069 | 0.0 | 0.0 | 0.0 | 457.865 | 3.166 | 0.0 | 36.054 |
| 19 | 360.468 | 6.85 | 69.37 | 23.068 | 3.166 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 30.682 | 1.088 | 3.307 | 0.977 |
| 20 | 68.259 | 75.004 | 229.675 | 21.378 | 2.117 | 0.779 | 0.0 | 5.042 | 11.415 | 19.162 | 19.718 | 22.382 | 1.033 | 2.409 | 16.509 | 4.094 |
| 21 | 13.061 | 8.676 | 51.664 | 9.423 | 44.303 | 22.524 | 7.205 | 23.323 | 66.492 | 69.793 | 110.562 | 19.055 | 6.908 | 7.909 | 32.268 | 5.81 |
| 22 | 35.022 | 24.459 | 52.757 | 18.526 | 47.95 | 25.577 | 7.155 | 28.22 | 49.772 | 54.577 | 56.986 | 40.365 | 5.085 | 15.14 | 23.419 | 13.967 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.057 | -0.329 | 0.601 | -1.023 | 0.798 | 0.246 | -0.843 | -0.04 | -0.191 | 0.173 | 0.214 | -0.319 | -1.574 | -0.413 | 0.626 | -0.61 |
| 02 | -0.621 | -0.515 | 0.697 | 0.055 | 0.553 | -0.256 | -1.176 | 0.027 | -0.149 | 0.787 | 0.4 | -0.209 | -1.367 | -0.36 | -0.085 | -0.329 |
| 03 | -1.214 | 0.272 | 0.213 | -0.607 | -0.519 | 0.868 | -1.493 | 0.063 | -1.638 | 1.041 | 0.007 | -0.368 | -1.67 | 0.79 | -0.074 | -1.221 |
| 04 | -3.167 | -1.392 | -0.984 | -0.494 | 0.02 | 0.086 | -1.991 | 1.869 | -1.594 | 0.071 | -1.237 | 0.605 | -1.716 | -0.306 | 0.274 | -1.006 |
| 05 | -4.398 | -1.616 | -0.064 | -1.218 | -2.207 | -2.104 | -1.552 | 0.995 | -1.713 | -1.464 | -0.35 | -0.001 | -1.699 | -1.897 | 2.147 | -0.916 |
| 06 | -2.448 | -2.645 | -1.45 | -2.369 | -1.548 | -1.832 | -3.176 | -1.247 | 0.024 | 1.004 | 1.325 | 1.064 | -2.938 | -1.574 | 1.303 | -0.775 |
| 07 | -1.108 | -1.366 | -0.302 | -2.959 | -0.078 | -0.323 | -1.856 | 0.309 | -0.091 | 0.855 | 0.851 | 0.747 | -0.628 | -0.349 | 0.383 | 0.033 |
| 08 | -0.509 | -0.432 | 0.172 | -1.334 | 0.507 | -0.514 | -0.935 | 0.327 | 0.164 | 0.559 | 0.129 | -0.449 | -0.592 | -0.023 | 0.642 | 0.119 |
| 09 | 0.056 | -0.402 | 0.683 | -1.256 | 0.217 | 0.168 | -1.089 | 0.192 | -0.297 | 0.186 | 0.653 | -0.287 | -1.275 | 0.025 | 0.128 | -0.027 |
| 10 | -0.63 | -0.544 | 0.557 | -0.767 | -0.072 | 0.165 | -1.579 | 0.569 | 0.038 | 0.455 | 0.607 | -0.08 | -1.35 | -0.223 | 0.323 | -0.25 |
| 11 | -0.787 | -0.25 | 0.155 | -1.022 | 0.223 | 0.434 | -1.336 | 0.076 | 0.195 | 0.316 | 0.516 | -0.098 | -0.906 | 0.22 | 0.201 | 0.058 |
| 12 | -0.052 | -0.791 | 0.523 | -1.446 | 0.656 | -0.109 | -1.123 | 0.583 | 0.211 | -0.539 | 0.605 | -0.388 | -0.367 | -0.781 | 0.532 | -0.591 |
| 13 | -0.113 | 0.729 | 0.356 | -0.892 | -1.398 | 0.07 | -0.929 | -0.39 | -0.645 | 1.011 | 0.569 | -0.677 | -1.489 | 0.099 | 0.286 | -0.512 |
| 14 | 0.228 | -2.294 | -4.398 | -0.651 | 1.359 | -1.434 | -3.251 | 1.039 | 1.204 | -1.854 | -1.925 | 0.248 | 0.489 | -4.398 | -2.954 | -0.695 |
| 15 | -4.398 | -4.398 | 2.31 | -4.398 | -4.398 | -4.398 | -0.753 | -4.398 | -4.398 | -4.398 | -1.55 | -4.398 | -4.398 | -4.398 | 1.628 | -4.398 |
| 16 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | 2.759 | -4.398 | -3.206 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 |
| 17 | -4.398 | -3.261 | -3.076 | 2.755 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -3.206 | -4.398 | -4.398 | -4.398 | -4.398 |
| 18 | -4.398 | -4.398 | -4.398 | -4.398 | -3.261 | -4.398 | -4.398 | -4.398 | -3.076 | -4.398 | -4.398 | -4.398 | 2.675 | -2.184 | -4.398 | 0.143 |
| 19 | 2.436 | -1.473 | 0.793 | -0.297 | -2.184 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -0.016 | -3.063 | -2.145 | -3.141 |
| 20 | 0.777 | 0.87 | 1.986 | -0.372 | -2.534 | -3.298 | -4.398 | -1.76 | -0.984 | -0.479 | -0.451 | -0.327 | -3.101 | -2.424 | -0.625 | -1.952 |
| 21 | -0.853 | -1.248 | 0.5 | -1.169 | 0.347 | -0.321 | -1.425 | -0.286 | 0.751 | 0.799 | 1.257 | -0.485 | -1.465 | -1.336 | 0.034 | -1.628 |
| 22 | 0.115 | -0.24 | 0.521 | -0.512 | 0.426 | -0.196 | -1.432 | -0.099 | 0.463 | 0.554 | 0.597 | 0.255 | -1.753 | -0.71 | -0.282 | -0.788 |