Model info
| Transcription factor | STAT1 | ||||||||
| Model | STAT1_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 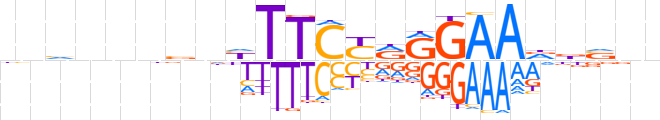 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Consensus | nnbvhTTCYYRGAAdhbnnnbnn | ||||||||
| wAUC | 0.9087851660828666 | ||||||||
| Best AUC | 0.9682459817874061 | ||||||||
| Benchmark datasets | 6 | ||||||||
| Aligned words | 498 | ||||||||
| TF family | STAT factors{6.2.1} | ||||||||
| TF subfamily | STAT1{6.2.1.0.1} | ||||||||
| HGNC | 11362 | ||||||||
| EntrezGene | 6772 | ||||||||
| UniProt ID | STAT1_HUMAN | ||||||||
| UniProt AC | P42224 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 50.875 | 23.632 | 33.101 | 46.271 | 13.109 | 68.651 | 4.767 | 44.508 | 8.893 | 44.628 | 33.988 | 18.62 | 12.869 | 40.276 | 36.18 | 11.631 |
| 02 | 5.683 | 30.127 | 29.711 | 20.227 | 14.716 | 116.659 | 13.094 | 32.718 | 11.736 | 58.344 | 19.598 | 18.357 | 2.89 | 63.338 | 43.048 | 11.753 |
| 03 | 12.788 | 2.718 | 13.689 | 5.831 | 139.384 | 76.106 | 18.199 | 34.778 | 47.0 | 36.564 | 20.624 | 1.264 | 21.049 | 25.267 | 28.275 | 8.465 |
| 04 | 32.861 | 36.145 | 23.566 | 127.649 | 29.832 | 32.324 | 0.927 | 77.572 | 10.229 | 30.286 | 8.905 | 31.366 | 6.763 | 11.082 | 7.736 | 24.757 |
| 05 | 1.156 | 0.0 | 1.953 | 76.576 | 0.0 | 0.0 | 0.0 | 109.837 | 0.92 | 1.168 | 0.0 | 39.046 | 0.0 | 1.053 | 2.171 | 258.121 |
| 06 | 0.0 | 0.0 | 2.076 | 0.0 | 0.0 | 0.0 | 1.053 | 1.168 | 0.0 | 0.0 | 2.078 | 2.046 | 1.988 | 0.0 | 29.379 | 452.213 |
| 07 | 0.0 | 1.988 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 29.463 | 3.967 | 0.0 | 1.156 | 24.444 | 428.106 | 0.0 | 2.877 |
| 08 | 28.866 | 20.099 | 0.945 | 3.997 | 5.714 | 345.575 | 1.003 | 81.769 | 0.0 | 0.0 | 0.0 | 0.0 | 2.067 | 1.966 | 0.0 | 0.0 |
| 09 | 6.851 | 5.419 | 1.871 | 22.506 | 37.92 | 190.287 | 4.156 | 135.276 | 0.945 | 0.0 | 1.003 | 0.0 | 6.127 | 57.981 | 5.314 | 16.344 |
| 10 | 6.707 | 0.0 | 45.135 | 0.0 | 156.162 | 8.669 | 85.803 | 3.051 | 10.504 | 0.904 | 0.0 | 0.936 | 27.639 | 2.827 | 139.011 | 4.651 |
| 11 | 2.049 | 0.0 | 185.61 | 13.352 | 3.008 | 0.966 | 6.354 | 2.072 | 8.812 | 2.921 | 225.011 | 33.205 | 0.0 | 0.0 | 8.638 | 0.0 |
| 12 | 12.935 | 0.934 | 0.0 | 0.0 | 1.833 | 2.054 | 0.0 | 0.0 | 390.822 | 21.977 | 7.033 | 5.782 | 46.728 | 1.901 | 0.0 | 0.0 |
| 13 | 445.631 | 0.894 | 4.88 | 0.914 | 25.894 | 0.0 | 0.0 | 0.972 | 5.929 | 1.105 | 0.0 | 0.0 | 4.729 | 0.0 | 1.053 | 0.0 |
| 14 | 269.919 | 38.75 | 89.587 | 83.927 | 0.0 | 0.0 | 1.105 | 0.894 | 1.903 | 0.965 | 3.066 | 0.0 | 0.914 | 0.0 | 0.972 | 0.0 |
| 15 | 72.192 | 79.415 | 37.493 | 83.636 | 10.541 | 10.076 | 0.94 | 18.157 | 15.667 | 29.052 | 14.849 | 35.161 | 11.451 | 36.855 | 6.183 | 30.331 |
| 16 | 11.436 | 61.908 | 23.743 | 12.764 | 28.071 | 58.33 | 10.238 | 58.759 | 2.693 | 26.722 | 19.913 | 10.137 | 17.13 | 72.788 | 58.813 | 18.554 |
| 17 | 18.323 | 18.614 | 19.372 | 3.022 | 98.367 | 44.454 | 8.684 | 68.243 | 42.321 | 22.371 | 35.889 | 12.126 | 34.841 | 13.155 | 28.483 | 23.736 |
| 18 | 43.058 | 75.888 | 47.107 | 27.799 | 34.113 | 33.253 | 9.549 | 21.679 | 22.865 | 24.734 | 27.081 | 17.749 | 11.142 | 51.436 | 27.386 | 17.161 |
| 19 | 15.926 | 9.672 | 48.783 | 36.798 | 63.081 | 34.466 | 27.523 | 60.241 | 27.357 | 27.096 | 40.061 | 16.609 | 3.725 | 20.286 | 47.184 | 13.191 |
| 20 | 14.46 | 13.913 | 24.378 | 57.338 | 20.681 | 18.796 | 15.525 | 36.518 | 23.962 | 27.729 | 46.348 | 65.513 | 6.001 | 20.865 | 47.31 | 52.663 |
| 21 | 7.641 | 14.827 | 29.131 | 13.505 | 21.606 | 28.758 | 7.524 | 23.415 | 23.746 | 22.505 | 59.613 | 27.698 | 19.085 | 38.199 | 88.794 | 65.953 |
| 22 | 32.304 | 12.714 | 18.354 | 8.707 | 36.973 | 29.792 | 4.919 | 32.605 | 106.231 | 27.168 | 33.2 | 18.463 | 16.229 | 21.31 | 65.028 | 28.003 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.499 | -0.26 | 0.073 | 0.404 | -0.836 | 0.796 | -1.799 | 0.366 | -1.21 | 0.369 | 0.099 | -0.494 | -0.854 | 0.267 | 0.161 | -0.952 |
| 02 | -1.635 | -0.02 | -0.034 | -0.412 | -0.723 | 1.324 | -0.837 | 0.061 | -0.943 | 0.635 | -0.443 | -0.508 | -2.251 | 0.716 | 0.333 | -0.942 |
| 03 | -0.86 | -2.305 | -0.794 | -1.611 | 1.502 | 0.899 | -0.516 | 0.122 | 0.42 | 0.171 | -0.393 | -2.937 | -0.373 | -0.194 | -0.083 | -1.258 |
| 04 | 0.066 | 0.16 | -0.262 | 1.414 | -0.03 | 0.049 | -3.165 | 0.918 | -1.076 | -0.015 | -1.209 | 0.02 | -1.471 | -0.999 | -1.344 | -0.214 |
| 05 | -3.005 | -4.387 | -2.588 | 0.905 | -4.387 | -4.387 | -4.387 | 1.264 | -3.17 | -2.997 | -4.387 | 0.236 | -4.387 | -3.074 | -2.499 | 2.117 |
| 06 | -4.387 | -4.387 | -2.537 | -4.387 | -4.387 | -4.387 | -3.074 | -2.997 | -4.387 | -4.387 | -2.536 | -2.549 | -2.573 | -4.387 | -0.045 | 2.677 |
| 07 | -4.387 | -2.573 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 | -4.387 | -0.042 | -1.967 | -4.387 | -3.005 | -0.226 | 2.622 | -4.387 | -2.255 |
| 08 | -0.062 | -0.419 | -3.151 | -1.96 | -1.63 | 2.408 | -3.109 | 0.97 | -4.387 | -4.387 | -4.387 | -4.387 | -2.541 | -2.583 | -4.387 | -4.387 |
| 09 | -1.459 | -1.68 | -2.624 | -0.308 | 0.207 | 1.812 | -1.925 | 1.472 | -3.151 | -4.387 | -3.109 | -4.387 | -1.564 | 0.628 | -1.698 | -0.621 |
| 10 | -1.479 | -4.387 | 0.38 | -4.387 | 1.615 | -1.235 | 1.018 | -2.203 | -1.05 | -3.183 | -4.387 | -3.158 | -0.105 | -2.271 | 1.499 | -1.821 |
| 11 | -2.548 | -4.387 | 1.787 | -0.818 | -2.216 | -3.136 | -1.53 | -2.539 | -1.219 | -2.242 | 1.979 | 0.076 | -4.387 | -4.387 | -1.238 | -4.387 |
| 12 | -0.849 | -3.16 | -4.387 | -4.387 | -2.641 | -2.546 | -4.387 | -4.387 | 2.531 | -0.331 | -1.434 | -1.619 | 0.414 | -2.611 | -4.387 | -4.387 |
| 13 | 2.662 | -3.191 | -1.777 | -3.175 | -0.17 | -4.387 | -4.387 | -3.132 | -1.595 | -3.038 | -4.387 | -4.387 | -1.806 | -4.387 | -3.073 | -4.387 |
| 14 | 2.161 | 0.229 | 1.061 | 0.996 | -4.387 | -4.387 | -3.038 | -3.191 | -2.61 | -3.137 | -2.199 | -4.387 | -3.175 | -4.387 | -3.132 | -4.387 |
| 15 | 0.846 | 0.941 | 0.196 | 0.993 | -1.047 | -1.091 | -3.155 | -0.518 | -0.662 | -0.056 | -0.715 | 0.132 | -0.967 | 0.179 | -1.556 | -0.014 |
| 16 | -0.968 | 0.693 | -0.255 | -0.862 | -0.09 | 0.634 | -1.075 | 0.642 | -2.313 | -0.139 | -0.428 | -1.085 | -0.575 | 0.854 | 0.643 | -0.497 |
| 17 | -0.509 | -0.494 | -0.455 | -2.212 | 1.154 | 0.365 | -1.233 | 0.79 | 0.316 | -0.313 | 0.153 | -0.912 | 0.123 | -0.833 | -0.076 | -0.255 |
| 18 | 0.333 | 0.896 | 0.422 | -0.1 | 0.103 | 0.077 | -1.142 | -0.344 | -0.292 | -0.215 | -0.125 | -0.541 | -0.994 | 0.509 | -0.114 | -0.573 |
| 19 | -0.646 | -1.13 | 0.457 | 0.177 | 0.712 | 0.113 | -0.109 | 0.666 | -0.115 | -0.125 | 0.262 | -0.605 | -2.024 | -0.41 | 0.424 | -0.83 |
| 20 | -0.741 | -0.778 | -0.229 | 0.617 | -0.391 | -0.484 | -0.671 | 0.17 | -0.246 | -0.102 | 0.406 | 0.75 | -1.584 | -0.382 | 0.426 | 0.533 |
| 21 | -1.355 | -0.716 | -0.053 | -0.807 | -0.348 | -0.066 | -1.37 | -0.269 | -0.255 | -0.308 | 0.656 | -0.103 | -0.469 | 0.214 | 1.052 | 0.756 |
| 22 | 0.049 | -0.866 | -0.508 | -1.231 | 0.182 | -0.031 | -1.769 | 0.058 | 1.231 | -0.122 | 0.076 | -0.502 | -0.628 | -0.361 | 0.742 | -0.092 |