Model info
| Transcription factor | STAT5B | ||||||||
| Model | STA5B_HUMAN.H10DI.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 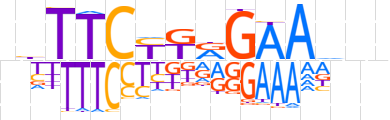 | ||||||||
| LOGO (reverse complement) | 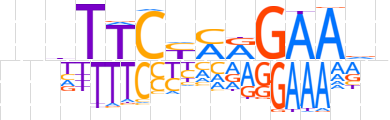 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 14 | ||||||||
| Quality | C | ||||||||
| Consensus | nbTTCYTRGAAdnn | ||||||||
| wAUC | 0.9217641911407325 | ||||||||
| Best AUC | 0.9217641911407325 | ||||||||
| Benchmark datasets | 1 | ||||||||
| Aligned words | 505 | ||||||||
| TF family | STAT factors{6.2.1} | ||||||||
| TF subfamily | STAT5B{6.2.1.0.6} | ||||||||
| HGNC | 11367 | ||||||||
| EntrezGene | 6777 | ||||||||
| UniProt ID | STA5B_HUMAN | ||||||||
| UniProt AC | P51692 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 33.359 | 29.656 | 36.093 | 49.376 | 16.652 | 29.941 | 6.963 | 43.282 | 14.774 | 36.949 | 17.876 | 61.249 | 6.897 | 30.768 | 20.436 | 64.728 |
| 02 | 0.0 | 1.925 | 1.012 | 68.745 | 3.08 | 1.997 | 0.0 | 122.238 | 0.0 | 4.817 | 0.0 | 76.551 | 0.997 | 4.171 | 0.0 | 213.467 |
| 03 | 0.0 | 0.0 | 1.057 | 3.02 | 1.039 | 0.0 | 0.0 | 11.871 | 0.0 | 0.0 | 0.0 | 1.012 | 26.915 | 0.957 | 2.835 | 450.294 |
| 04 | 0.884 | 27.07 | 0.0 | 0.0 | 0.957 | 0.0 | 0.0 | 0.0 | 0.0 | 3.891 | 0.0 | 0.0 | 4.083 | 455.155 | 0.0 | 6.959 |
| 05 | 0.0 | 2.086 | 0.0 | 3.839 | 31.168 | 140.386 | 5.986 | 308.576 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.867 | 0.0 | 2.091 |
| 06 | 0.0 | 0.0 | 26.232 | 4.936 | 0.0 | 0.0 | 11.687 | 135.652 | 0.0 | 0.0 | 2.918 | 3.068 | 0.0 | 0.0 | 201.152 | 113.355 |
| 07 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 161.797 | 4.033 | 53.44 | 22.718 | 84.931 | 5.838 | 150.144 | 16.099 |
| 08 | 0.0 | 0.0 | 240.615 | 6.113 | 0.0 | 0.0 | 8.948 | 0.923 | 4.039 | 0.988 | 188.509 | 10.047 | 0.0 | 1.059 | 37.759 | 0.0 |
| 09 | 4.039 | 0.0 | 0.0 | 0.0 | 2.047 | 0.0 | 0.0 | 0.0 | 416.758 | 14.62 | 11.93 | 32.522 | 16.054 | 1.03 | 0.0 | 0.0 |
| 10 | 432.093 | 0.0 | 5.832 | 0.973 | 15.65 | 0.0 | 0.0 | 0.0 | 11.93 | 0.0 | 0.0 | 0.0 | 30.689 | 0.0 | 1.833 | 0.0 |
| 11 | 208.816 | 87.21 | 100.463 | 93.873 | 0.0 | 0.0 | 0.0 | 0.0 | 0.902 | 1.826 | 2.993 | 1.944 | 0.0 | 0.0 | 0.0 | 0.973 |
| 12 | 53.182 | 50.671 | 33.537 | 72.329 | 19.87 | 21.52 | 3.793 | 43.854 | 24.654 | 31.753 | 20.884 | 26.164 | 9.123 | 22.267 | 27.158 | 38.242 |
| 13 | 32.231 | 19.819 | 44.71 | 10.069 | 38.335 | 45.711 | 3.886 | 38.279 | 23.133 | 29.324 | 22.019 | 10.896 | 30.416 | 37.852 | 71.981 | 40.34 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.067 | -0.05 | 0.144 | 0.455 | -0.617 | -0.04 | -1.457 | 0.324 | -0.734 | 0.168 | -0.547 | 0.669 | -1.466 | -0.013 | -0.416 | 0.724 |
| 02 | -4.398 | -2.614 | -3.116 | 0.784 | -2.209 | -2.583 | -4.398 | 1.357 | -4.398 | -1.803 | -4.398 | 0.891 | -3.127 | -1.935 | -4.398 | 1.913 |
| 03 | -4.398 | -4.398 | -3.084 | -2.226 | -3.097 | -4.398 | -4.398 | -0.946 | -4.398 | -4.398 | -4.398 | -3.116 | -0.145 | -3.156 | -2.282 | 2.658 |
| 04 | -3.211 | -0.14 | -4.398 | -4.398 | -3.156 | -4.398 | -4.398 | -4.398 | -4.398 | -1.999 | -4.398 | -4.398 | -1.955 | 2.669 | -4.398 | -1.458 |
| 05 | -4.398 | -2.546 | -4.398 | -2.011 | -0.001 | 1.495 | -1.6 | 2.281 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -1.793 | -4.398 | -2.544 |
| 06 | -4.398 | -4.398 | -0.171 | -1.78 | -4.398 | -4.398 | -0.961 | 1.461 | -4.398 | -4.398 | -2.257 | -2.212 | -4.398 | -4.398 | 1.854 | 1.282 |
| 07 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | 1.636 | -1.966 | 0.533 | -0.312 | 0.994 | -1.624 | 1.562 | -0.65 |
| 08 | -4.398 | -4.398 | 2.032 | -1.58 | -4.398 | -4.398 | -1.218 | -3.181 | -1.965 | -3.133 | 1.789 | -1.107 | -4.398 | -3.083 | 0.189 | -4.398 |
| 09 | -1.965 | -4.398 | -4.398 | -4.398 | -2.562 | -4.398 | -4.398 | -4.398 | 2.581 | -0.744 | -0.941 | 0.041 | -0.653 | -3.103 | -4.398 | -4.398 |
| 10 | 2.617 | -4.398 | -1.625 | -3.144 | -0.677 | -4.398 | -4.398 | -4.398 | -0.941 | -4.398 | -4.398 | -4.398 | -0.016 | -4.398 | -2.654 | -4.398 |
| 11 | 1.891 | 1.02 | 1.161 | 1.094 | -4.398 | -4.398 | -4.398 | -4.398 | -3.197 | -2.658 | -2.234 | -2.606 | -4.398 | -4.398 | -4.398 | -3.144 |
| 12 | 0.529 | 0.481 | 0.072 | 0.834 | -0.444 | -0.366 | -2.022 | 0.337 | -0.232 | 0.018 | -0.395 | -0.173 | -1.2 | -0.332 | -0.137 | 0.202 |
| 13 | 0.033 | -0.446 | 0.356 | -1.105 | 0.204 | 0.378 | -2.0 | 0.203 | -0.294 | -0.061 | -0.343 | -1.029 | -0.025 | 0.192 | 0.829 | 0.255 |