Model info
| Transcription factor | STAT5A | ||||||||
| Model | STA5A_HUMAN.H10DI.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 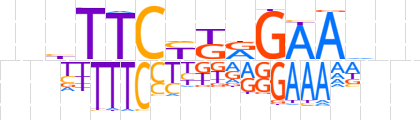 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 15 | ||||||||
| Quality | B | ||||||||
| Consensus | nnbTTCYGRGAAdnn | ||||||||
| wAUC | 0.835154785981265 | ||||||||
| Best AUC | 0.9047646436786899 | ||||||||
| Benchmark datasets | 2 | ||||||||
| Aligned words | 498 | ||||||||
| TF family | STAT factors{6.2.1} | ||||||||
| TF subfamily | STAT5A{6.2.1.0.5} | ||||||||
| HGNC | 11366 | ||||||||
| EntrezGene | 6776 | ||||||||
| UniProt ID | STA5A_HUMAN | ||||||||
| UniProt AC | P42229 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 30.667 | 12.831 | 47.332 | 8.852 | 68.351 | 20.912 | 19.704 | 58.509 | 25.679 | 24.75 | 32.928 | 22.14 | 25.475 | 15.921 | 46.713 | 31.979 |
| 02 | 21.198 | 29.792 | 33.955 | 65.226 | 10.463 | 22.977 | 4.864 | 36.109 | 24.089 | 45.968 | 15.646 | 60.975 | 8.784 | 30.245 | 12.523 | 69.929 |
| 03 | 0.0 | 2.09 | 0.0 | 62.444 | 1.889 | 4.056 | 0.888 | 122.149 | 0.0 | 0.951 | 0.994 | 65.044 | 0.0 | 5.677 | 0.0 | 226.561 |
| 04 | 0.0 | 0.0 | 0.0 | 1.889 | 0.992 | 0.0 | 0.0 | 11.781 | 0.0 | 0.0 | 0.0 | 1.882 | 21.711 | 0.926 | 4.844 | 448.718 |
| 05 | 0.0 | 22.703 | 0.0 | 0.0 | 0.0 | 0.926 | 0.0 | 0.0 | 0.0 | 4.844 | 0.0 | 0.0 | 6.041 | 452.084 | 1.008 | 5.138 |
| 06 | 2.065 | 3.053 | 0.0 | 0.923 | 30.783 | 151.487 | 13.231 | 285.056 | 0.0 | 0.0 | 1.008 | 0.0 | 0.0 | 2.941 | 1.053 | 1.144 |
| 07 | 0.0 | 0.0 | 23.601 | 9.247 | 0.0 | 0.0 | 26.33 | 131.151 | 0.0 | 0.0 | 7.017 | 8.275 | 0.0 | 0.0 | 195.68 | 91.443 |
| 08 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 166.156 | 5.079 | 60.907 | 20.485 | 71.529 | 3.063 | 153.852 | 11.671 |
| 09 | 1.954 | 1.859 | 225.929 | 7.944 | 1.802 | 0.0 | 6.34 | 0.0 | 0.0 | 1.921 | 204.875 | 7.963 | 0.0 | 0.0 | 32.156 | 0.0 |
| 10 | 1.954 | 0.913 | 0.888 | 0.0 | 1.947 | 0.929 | 0.0 | 0.903 | 413.291 | 16.132 | 3.302 | 36.576 | 13.789 | 2.118 | 0.0 | 0.0 |
| 11 | 413.036 | 1.048 | 14.692 | 2.206 | 20.093 | 0.0 | 0.0 | 0.0 | 3.131 | 0.0 | 1.059 | 0.0 | 31.68 | 3.876 | 0.998 | 0.924 |
| 12 | 194.347 | 60.076 | 98.094 | 115.422 | 0.0 | 0.0 | 2.966 | 1.958 | 6.99 | 3.826 | 1.028 | 4.905 | 0.0 | 0.0 | 1.893 | 1.238 |
| 13 | 55.485 | 48.4 | 33.258 | 64.193 | 13.473 | 16.744 | 0.947 | 32.739 | 29.741 | 30.948 | 14.777 | 28.515 | 11.869 | 32.908 | 33.328 | 45.418 |
| 14 | 25.76 | 19.977 | 45.678 | 19.152 | 43.791 | 33.089 | 6.106 | 46.014 | 20.573 | 33.933 | 19.985 | 7.818 | 19.089 | 45.193 | 64.962 | 41.621 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.004 | -0.858 | 0.425 | -1.216 | 0.79 | -0.381 | -0.44 | 0.636 | -0.179 | -0.216 | 0.066 | -0.325 | -0.187 | -0.648 | 0.412 | 0.037 |
| 02 | -0.368 | -0.033 | 0.096 | 0.744 | -1.056 | -0.289 | -1.781 | 0.157 | -0.242 | 0.396 | -0.665 | 0.677 | -1.224 | -0.018 | -0.882 | 0.813 |
| 03 | -4.388 | -2.533 | -4.388 | 0.701 | -2.617 | -1.949 | -3.196 | 1.369 | -4.388 | -3.148 | -3.117 | 0.741 | -4.388 | -1.637 | -4.388 | 1.985 |
| 04 | -4.388 | -4.388 | -4.388 | -2.617 | -3.118 | -4.388 | -4.388 | -0.941 | -4.388 | -4.388 | -4.388 | -2.62 | -0.344 | -3.168 | -1.785 | 2.667 |
| 05 | -4.388 | -0.301 | -4.388 | -4.388 | -4.388 | -3.168 | -4.388 | -4.388 | -4.388 | -1.785 | -4.388 | -4.388 | -1.579 | 2.675 | -3.107 | -1.731 |
| 06 | -2.543 | -2.204 | -4.388 | -3.17 | -0.0 | 1.583 | -0.829 | 2.214 | -4.388 | -4.388 | -3.107 | -4.388 | -4.388 | -2.237 | -3.075 | -3.013 |
| 07 | -4.388 | -4.388 | -0.262 | -1.175 | -4.388 | -4.388 | -0.155 | 1.439 | -4.388 | -4.388 | -1.438 | -1.281 | -4.388 | -4.388 | 1.839 | 1.08 |
| 08 | -4.388 | -4.388 | -4.388 | -4.388 | -4.388 | -4.388 | -4.388 | -4.388 | 1.675 | -1.741 | 0.676 | -0.401 | 0.836 | -2.201 | 1.599 | -0.95 |
| 09 | -2.589 | -2.63 | 1.982 | -1.32 | -2.656 | -4.388 | -1.534 | -4.388 | -4.388 | -2.603 | 1.884 | -1.318 | -4.388 | -4.388 | 0.043 | -4.388 |
| 10 | -2.589 | -3.177 | -3.196 | -4.388 | -2.592 | -3.165 | -4.388 | -3.185 | 2.585 | -0.635 | -2.134 | 0.17 | -0.788 | -2.521 | -4.388 | -4.388 |
| 11 | 2.585 | -3.079 | -0.727 | -2.487 | -0.42 | -4.388 | -4.388 | -4.388 | -2.182 | -4.388 | -3.071 | -4.388 | 0.028 | -1.99 | -3.114 | -3.169 |
| 12 | 1.832 | 0.662 | 1.15 | 1.312 | -4.388 | -4.388 | -2.23 | -2.587 | -1.442 | -2.002 | -3.093 | -1.774 | -4.388 | -4.388 | -2.616 | -2.954 |
| 13 | 0.583 | 0.448 | 0.076 | 0.728 | -0.811 | -0.599 | -3.152 | 0.06 | -0.034 | 0.005 | -0.721 | -0.076 | -0.934 | 0.066 | 0.078 | 0.385 |
| 14 | -0.176 | -0.426 | 0.39 | -0.467 | 0.348 | 0.071 | -1.569 | 0.397 | -0.397 | 0.096 | -0.426 | -1.335 | -0.471 | 0.38 | 0.74 | 0.298 |