Model info
| Transcription factor | RUNX3 | ||||||||
| Model | RUNX3_HUMAN.H10DI.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 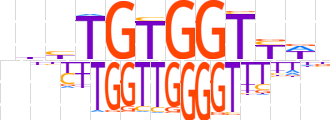 | ||||||||
| LOGO (reverse complement) | 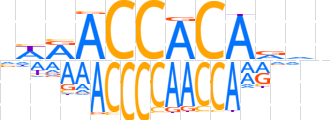 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 12 | ||||||||
| Quality | C | ||||||||
| Consensus | nbbTGTGGTYWn | ||||||||
| wAUC | 0.9119485630909621 | ||||||||
| Best AUC | 0.9119485630909621 | ||||||||
| Benchmark datasets | 1 | ||||||||
| Aligned words | 516 | ||||||||
| TF family | Runt-related factors{6.4.1} | ||||||||
| TF subfamily | Runx3 (PEBP2alphaC, CBF-alpha3, AML-2){6.4.1.0.3} | ||||||||
| HGNC | 10473 | ||||||||
| EntrezGene | 864 | ||||||||
| UniProt ID | RUNX3_HUMAN | ||||||||
| UniProt AC | Q13761 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 14.195 | 18.767 | 33.484 | 32.15 | 29.86 | 21.644 | 3.865 | 54.433 | 10.894 | 22.063 | 44.416 | 62.027 | 16.225 | 35.958 | 26.81 | 73.208 |
| 02 | 7.669 | 36.242 | 15.07 | 12.194 | 10.04 | 39.674 | 3.845 | 44.873 | 18.801 | 44.523 | 4.934 | 40.318 | 5.904 | 83.583 | 21.78 | 110.551 |
| 03 | 6.866 | 0.0 | 1.032 | 34.515 | 6.858 | 4.06 | 2.058 | 191.047 | 2.063 | 0.0 | 0.0 | 43.566 | 3.979 | 4.212 | 4.027 | 195.716 |
| 04 | 0.0 | 0.0 | 19.766 | 0.0 | 0.0 | 0.0 | 8.272 | 0.0 | 0.0 | 0.0 | 7.118 | 0.0 | 0.0 | 0.953 | 460.993 | 2.898 |
| 05 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.953 | 9.993 | 45.836 | 0.0 | 440.321 | 0.0 | 0.0 | 0.0 | 2.898 |
| 06 | 0.0 | 0.0 | 9.993 | 0.0 | 0.0 | 0.0 | 45.836 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.024 | 0.0 | 443.147 | 0.0 |
| 07 | 0.0 | 0.0 | 1.024 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 497.026 | 1.949 | 0.0 | 0.0 | 0.0 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 22.004 | 10.866 | 465.18 | 0.0 | 0.0 | 0.0 | 1.949 |
| 09 | 0.0 | 0.0 | 0.0 | 0.0 | 1.037 | 1.973 | 0.0 | 18.994 | 0.922 | 1.956 | 0.0 | 7.988 | 16.93 | 89.602 | 43.907 | 316.69 |
| 10 | 5.85 | 0.983 | 6.187 | 5.869 | 53.611 | 2.063 | 1.994 | 35.863 | 19.949 | 0.956 | 4.985 | 18.017 | 88.478 | 8.888 | 38.701 | 207.605 |
| 11 | 43.83 | 28.479 | 71.113 | 24.467 | 8.962 | 0.962 | 0.962 | 2.004 | 12.903 | 14.221 | 10.808 | 13.935 | 36.131 | 63.342 | 74.872 | 93.008 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.774 | -0.502 | 0.068 | 0.028 | -0.045 | -0.362 | -2.007 | 0.55 | -1.031 | -0.343 | 0.348 | 0.679 | -0.644 | 0.139 | -0.151 | 0.844 |
| 02 | -1.368 | 0.147 | -0.716 | -0.922 | -1.11 | 0.236 | -2.011 | 0.358 | -0.5 | 0.35 | -1.782 | 0.252 | -1.615 | 0.976 | -0.356 | 1.255 |
| 03 | -1.473 | -4.4 | -3.103 | 0.098 | -1.474 | -1.962 | -2.56 | 1.8 | -2.558 | -4.4 | -4.4 | 0.329 | -1.98 | -1.928 | -1.969 | 1.824 |
| 04 | -4.4 | -4.4 | -0.451 | -4.4 | -4.4 | -4.4 | -1.296 | -4.4 | -4.4 | -4.4 | -1.439 | -4.4 | -4.4 | -3.161 | 2.68 | -2.265 |
| 05 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -3.161 | -1.114 | 0.379 | -4.4 | 2.634 | -4.4 | -4.4 | -4.4 | -2.265 |
| 06 | -4.4 | -4.4 | -1.114 | -4.4 | -4.4 | -4.4 | 0.379 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -3.109 | -4.4 | 2.64 | -4.4 |
| 07 | -4.4 | -4.4 | -3.109 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | 2.755 | -2.605 | -4.4 | -4.4 | -4.4 | -4.4 |
| 08 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -0.346 | -1.034 | 2.689 | -4.4 | -4.4 | -4.4 | -2.605 |
| 09 | -4.4 | -4.4 | -4.4 | -4.4 | -3.1 | -2.595 | -4.4 | -0.49 | -3.184 | -2.602 | -4.4 | -1.329 | -0.603 | 1.045 | 0.337 | 2.305 |
| 10 | -1.624 | -3.138 | -1.571 | -1.621 | 0.535 | -2.558 | -2.586 | 0.136 | -0.442 | -3.158 | -1.773 | -0.542 | 1.033 | -1.227 | 0.211 | 1.883 |
| 11 | 0.335 | -0.092 | 0.815 | -0.241 | -1.219 | -3.154 | -3.154 | -2.582 | -0.867 | -0.773 | -1.039 | -0.793 | 0.143 | 0.7 | 0.867 | 1.082 |