Model info
| Transcription factor | Pou5f1 | ||||||||
| Model | PO5F1_MOUSE.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 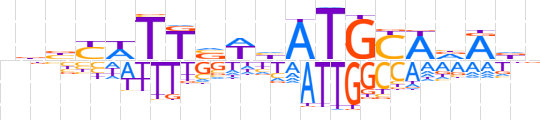 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 19 | ||||||||
| Quality | A | ||||||||
| Consensus | nbbYWTTvWhATGCAdRdn | ||||||||
| wAUC | 0.902146413845935 | ||||||||
| Best AUC | 0.9725630727094943 | ||||||||
| Benchmark datasets | 6 | ||||||||
| Aligned words | 505 | ||||||||
| TF family | POU domain factors{3.1.10} | ||||||||
| TF subfamily | POU5 (Oct-3/4-like factors){3.1.10.5} | ||||||||
| MGI | 101893 | ||||||||
| EntrezGene | 18999 | ||||||||
| UniProt ID | PO5F1_MOUSE | ||||||||
| UniProt AC | P20263 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 20.317 | 17.744 | 42.945 | 27.405 | 20.647 | 27.2 | 7.193 | 73.898 | 8.964 | 33.267 | 47.909 | 43.486 | 4.06 | 38.053 | 37.775 | 47.319 |
| 02 | 2.727 | 20.742 | 21.401 | 9.119 | 13.576 | 40.55 | 11.577 | 50.56 | 6.139 | 80.984 | 18.809 | 29.891 | 7.928 | 85.972 | 46.177 | 52.031 |
| 03 | 0.0 | 17.394 | 2.207 | 10.768 | 10.134 | 128.164 | 2.472 | 87.477 | 6.776 | 58.443 | 7.734 | 25.012 | 3.88 | 100.531 | 9.911 | 27.279 |
| 04 | 13.637 | 1.91 | 3.954 | 1.29 | 161.391 | 12.948 | 3.005 | 127.186 | 15.081 | 0.0 | 1.019 | 6.225 | 65.304 | 7.761 | 6.621 | 70.852 |
| 05 | 2.101 | 3.039 | 5.694 | 244.579 | 2.838 | 2.169 | 0.987 | 16.624 | 0.0 | 1.067 | 1.019 | 12.513 | 0.0 | 12.321 | 5.585 | 187.648 |
| 06 | 0.0 | 0.0 | 1.937 | 3.002 | 1.067 | 1.082 | 2.134 | 14.312 | 3.282 | 3.321 | 2.832 | 3.849 | 10.924 | 1.91 | 62.408 | 386.121 |
| 07 | 0.966 | 1.929 | 10.524 | 1.854 | 4.362 | 1.951 | 0.0 | 0.0 | 28.849 | 24.045 | 13.524 | 2.893 | 39.324 | 102.131 | 242.533 | 23.296 |
| 08 | 36.643 | 1.917 | 1.983 | 32.959 | 87.499 | 2.086 | 1.829 | 38.643 | 38.666 | 2.868 | 15.078 | 209.969 | 16.48 | 0.0 | 0.951 | 10.612 |
| 09 | 47.183 | 14.347 | 19.664 | 98.094 | 0.91 | 2.987 | 0.0 | 2.974 | 2.923 | 3.977 | 1.94 | 11.0 | 25.846 | 95.894 | 33.503 | 136.939 |
| 10 | 65.649 | 4.753 | 0.892 | 5.568 | 105.258 | 0.0 | 2.28 | 9.666 | 49.029 | 1.006 | 1.129 | 3.943 | 224.404 | 3.566 | 7.705 | 13.332 |
| 11 | 5.619 | 1.121 | 3.267 | 434.334 | 4.707 | 0.882 | 0.0 | 3.737 | 0.0 | 0.0 | 0.0 | 12.006 | 1.013 | 0.872 | 0.0 | 30.625 |
| 12 | 0.0 | 0.936 | 10.404 | 0.0 | 0.0 | 0.0 | 0.882 | 1.993 | 0.0 | 0.0 | 3.267 | 0.0 | 3.805 | 0.919 | 435.595 | 40.382 |
| 13 | 0.936 | 2.869 | 0.0 | 0.0 | 0.0 | 0.936 | 0.0 | 0.919 | 8.721 | 355.595 | 23.677 | 62.155 | 2.852 | 35.589 | 1.976 | 1.958 |
| 14 | 5.74 | 0.0 | 0.0 | 6.769 | 313.218 | 5.981 | 22.361 | 53.429 | 19.799 | 0.0 | 0.0 | 5.854 | 47.668 | 0.892 | 0.0 | 16.473 |
| 15 | 212.746 | 40.316 | 71.925 | 61.438 | 5.027 | 0.0 | 0.0 | 1.845 | 0.0 | 15.816 | 1.924 | 4.622 | 55.79 | 1.109 | 18.735 | 6.891 |
| 16 | 244.225 | 0.973 | 11.88 | 16.485 | 47.447 | 1.835 | 4.263 | 3.695 | 67.61 | 4.83 | 8.59 | 11.554 | 34.821 | 6.118 | 22.367 | 11.49 |
| 17 | 53.969 | 39.694 | 71.842 | 228.6 | 1.938 | 3.657 | 0.0 | 8.161 | 22.435 | 7.226 | 10.835 | 6.603 | 0.91 | 7.853 | 12.687 | 21.774 |
| 18 | 22.967 | 15.331 | 23.597 | 17.358 | 15.665 | 19.174 | 7.197 | 16.393 | 29.573 | 27.147 | 12.81 | 25.834 | 50.66 | 41.593 | 100.383 | 72.5 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.42 | -0.553 | 0.318 | -0.126 | -0.405 | -0.133 | -1.425 | 0.857 | -1.215 | 0.065 | 0.427 | 0.331 | -1.958 | 0.198 | 0.191 | 0.414 |
| 02 | -2.315 | -0.4 | -0.369 | -1.199 | -0.814 | 0.261 | -0.969 | 0.48 | -1.575 | 0.948 | -0.496 | -0.04 | -1.333 | 1.008 | 0.39 | 0.509 |
| 03 | -4.397 | -0.573 | -2.497 | -1.039 | -1.097 | 1.406 | -2.4 | 1.025 | -1.482 | 0.624 | -1.356 | -0.216 | -2.0 | 1.164 | -1.119 | -0.131 |
| 04 | -0.81 | -2.619 | -1.982 | -2.933 | 1.635 | -0.86 | -2.229 | 1.398 | -0.712 | -4.397 | -3.109 | -1.562 | 0.734 | -1.353 | -1.504 | 0.815 |
| 05 | -2.539 | -2.219 | -1.645 | 2.05 | -2.279 | -2.512 | -3.132 | -0.617 | -4.397 | -3.075 | -3.109 | -0.893 | -4.397 | -0.908 | -1.664 | 1.786 |
| 06 | -4.397 | -4.397 | -2.607 | -2.23 | -3.075 | -3.065 | -2.526 | -0.763 | -2.151 | -2.14 | -2.281 | -2.007 | -1.025 | -2.619 | 0.689 | 2.506 |
| 07 | -3.147 | -2.61 | -1.061 | -2.643 | -1.893 | -2.601 | -4.397 | -4.397 | -0.075 | -0.255 | -0.818 | -2.263 | 0.231 | 1.179 | 2.042 | -0.286 |
| 08 | 0.161 | -2.616 | -2.587 | 0.056 | 1.025 | -2.545 | -2.655 | 0.214 | 0.214 | -2.27 | -0.712 | 1.898 | -0.625 | -4.397 | -3.158 | -1.053 |
| 09 | 0.411 | -0.761 | -0.452 | 1.139 | -3.19 | -2.234 | -4.397 | -2.238 | -2.253 | -1.977 | -2.606 | -1.018 | -0.184 | 1.117 | 0.072 | 1.472 |
| 10 | 0.739 | -1.813 | -3.204 | -1.666 | 1.209 | -4.397 | -2.469 | -1.143 | 0.45 | -3.118 | -3.034 | -1.985 | 1.964 | -2.076 | -1.36 | -0.832 |
| 11 | -1.658 | -3.039 | -2.155 | 2.624 | -1.822 | -3.212 | -4.397 | -2.034 | -4.397 | -4.397 | -4.397 | -0.934 | -3.114 | -3.22 | -4.397 | -0.016 |
| 12 | -4.397 | -3.17 | -1.072 | -4.397 | -4.397 | -4.397 | -3.212 | -2.583 | -4.397 | -4.397 | -2.155 | -4.397 | -2.017 | -3.183 | 2.627 | 0.257 |
| 13 | -3.17 | -2.27 | -4.397 | -4.397 | -4.397 | -3.17 | -4.397 | -3.183 | -1.242 | 2.424 | -0.27 | 0.685 | -2.275 | 0.132 | -2.59 | -2.598 |
| 14 | -1.638 | -4.397 | -4.397 | -1.483 | 2.297 | -1.599 | -0.326 | 0.535 | -0.446 | -4.397 | -4.397 | -1.619 | 0.422 | -3.204 | -4.397 | -0.626 |
| 15 | 1.911 | 0.256 | 0.83 | 0.674 | -1.761 | -4.397 | -4.397 | -2.647 | -4.397 | -0.666 | -2.613 | -1.839 | 0.578 | -3.047 | -0.5 | -1.466 |
| 16 | 2.049 | -3.142 | -0.944 | -0.625 | 0.417 | -2.652 | -1.914 | -2.044 | 0.769 | -1.799 | -1.256 | -0.971 | 0.111 | -1.578 | -0.326 | -0.976 |
| 17 | 0.545 | 0.24 | 0.829 | 1.983 | -2.606 | -2.053 | -4.397 | -1.305 | -0.323 | -1.421 | -1.033 | -1.506 | -3.19 | -1.342 | -0.88 | -0.352 |
| 18 | -0.3 | -0.696 | -0.273 | -0.575 | -0.675 | -0.477 | -1.425 | -0.631 | -0.051 | -0.135 | -0.871 | -0.184 | 0.482 | 0.286 | 1.162 | 0.838 |