Model info
| Transcription factor | TP63 | ||||||||
| Model | P63_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 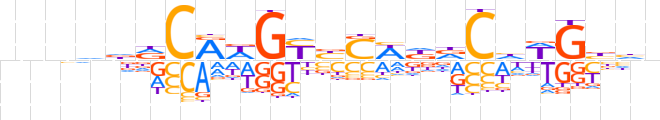 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Consensus | vdvCWdGYhWSvRCWYGYhbnnn | ||||||||
| wAUC | 0.9641803989928484 | ||||||||
| Best AUC | 0.9665663493487039 | ||||||||
| Benchmark datasets | 2 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | p53-related factors{6.3.1} | ||||||||
| TF subfamily | p63{6.3.1.0.2} | ||||||||
| HGNC | 15979 | ||||||||
| EntrezGene | 8626 | ||||||||
| UniProt ID | P63_HUMAN | ||||||||
| UniProt AC | Q9H3D4 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 31.572 | 4.899 | 63.956 | 69.24 | 24.449 | 4.835 | 9.82 | 38.041 | 52.994 | 19.615 | 53.541 | 37.152 | 7.779 | 4.794 | 34.625 | 25.723 |
| 02 | 59.963 | 2.94 | 45.904 | 7.988 | 30.211 | 1.0 | 1.044 | 1.888 | 69.827 | 5.004 | 82.179 | 4.933 | 27.675 | 65.132 | 63.646 | 13.702 |
| 03 | 10.44 | 171.43 | 3.893 | 1.913 | 49.17 | 23.964 | 0.0 | 0.943 | 2.011 | 189.799 | 0.0 | 0.963 | 0.0 | 27.438 | 1.073 | 0.0 |
| 04 | 5.654 | 1.162 | 0.945 | 53.86 | 312.181 | 40.029 | 21.406 | 39.015 | 1.073 | 2.917 | 0.0 | 0.976 | 0.0 | 0.963 | 1.904 | 0.952 |
| 05 | 136.03 | 3.002 | 68.527 | 111.349 | 6.765 | 1.005 | 0.0 | 37.301 | 5.811 | 0.0 | 1.933 | 16.511 | 10.885 | 30.254 | 10.064 | 43.6 |
| 06 | 25.528 | 0.995 | 132.969 | 0.0 | 0.0 | 0.0 | 34.261 | 0.0 | 0.0 | 0.0 | 79.524 | 0.999 | 0.957 | 0.0 | 207.803 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 26.485 | 0.995 | 0.0 | 0.0 | 0.0 | 114.907 | 159.873 | 1.899 | 177.879 | 0.999 | 0.0 | 0.0 | 0.0 |
| 08 | 67.355 | 37.54 | 0.979 | 11.027 | 29.761 | 100.839 | 1.008 | 28.265 | 0.0 | 1.899 | 0.0 | 0.0 | 54.153 | 88.792 | 10.704 | 50.714 |
| 09 | 9.942 | 5.829 | 30.744 | 104.754 | 60.155 | 4.801 | 7.867 | 156.248 | 2.938 | 0.0 | 1.926 | 7.827 | 13.517 | 1.105 | 41.969 | 33.416 |
| 10 | 2.988 | 8.616 | 71.881 | 3.067 | 0.0 | 0.996 | 10.738 | 0.0 | 10.82 | 9.81 | 58.889 | 2.987 | 20.708 | 43.676 | 227.251 | 10.61 |
| 11 | 3.874 | 10.655 | 19.987 | 0.0 | 26.155 | 32.069 | 2.878 | 1.996 | 58.014 | 46.041 | 230.264 | 34.44 | 3.004 | 0.0 | 12.703 | 0.957 |
| 12 | 55.707 | 0.975 | 24.52 | 9.845 | 82.932 | 0.96 | 1.055 | 3.817 | 148.32 | 2.945 | 94.004 | 20.562 | 9.959 | 0.973 | 24.53 | 1.933 |
| 13 | 21.502 | 271.141 | 1.148 | 3.127 | 1.933 | 3.921 | 0.0 | 0.0 | 0.0 | 144.109 | 0.0 | 0.0 | 0.0 | 36.157 | 0.0 | 0.0 |
| 14 | 3.38 | 1.941 | 0.0 | 18.113 | 191.743 | 53.807 | 8.785 | 200.993 | 0.0 | 1.148 | 0.0 | 0.0 | 2.192 | 0.0 | 0.0 | 0.935 |
| 15 | 12.862 | 25.796 | 24.453 | 134.205 | 1.0 | 3.849 | 0.0 | 52.046 | 0.98 | 0.0 | 0.981 | 6.824 | 9.861 | 31.749 | 5.543 | 172.888 |
| 16 | 0.0 | 0.0 | 24.703 | 0.0 | 2.883 | 1.105 | 56.463 | 0.943 | 0.0 | 0.0 | 30.976 | 0.0 | 5.5 | 0.0 | 360.463 | 0.0 |
| 17 | 2.086 | 3.414 | 0.0 | 2.883 | 0.0 | 1.105 | 0.0 | 0.0 | 102.581 | 204.549 | 2.912 | 162.563 | 0.0 | 0.943 | 0.0 | 0.0 |
| 18 | 82.195 | 11.749 | 3.921 | 6.802 | 69.387 | 87.438 | 4.883 | 48.303 | 0.946 | 0.961 | 1.006 | 0.0 | 23.63 | 61.784 | 29.173 | 50.859 |
| 19 | 15.767 | 30.038 | 54.847 | 75.505 | 33.299 | 44.133 | 6.749 | 77.751 | 3.846 | 10.66 | 9.817 | 14.66 | 16.641 | 37.062 | 28.299 | 23.962 |
| 20 | 10.955 | 9.951 | 29.174 | 19.473 | 50.008 | 31.065 | 9.696 | 31.124 | 44.037 | 18.61 | 22.378 | 14.686 | 12.927 | 25.772 | 120.114 | 33.066 |
| 21 | 11.849 | 29.81 | 34.511 | 41.757 | 34.48 | 15.505 | 4.967 | 30.446 | 26.638 | 33.531 | 96.712 | 24.481 | 9.923 | 28.191 | 33.898 | 26.337 |
| 22 | 28.138 | 14.681 | 31.204 | 8.867 | 28.243 | 25.247 | 3.852 | 49.697 | 88.815 | 25.672 | 38.053 | 17.549 | 11.76 | 23.435 | 40.418 | 47.408 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.044 | -1.755 | 0.744 | 0.823 | -0.208 | -1.768 | -1.097 | 0.229 | 0.557 | -0.424 | 0.567 | 0.205 | -1.32 | -1.775 | 0.135 | -0.158 |
| 02 | 0.68 | -2.218 | 0.415 | -1.295 | 0.001 | -3.093 | -3.063 | -2.599 | 0.831 | -1.736 | 0.993 | -1.749 | -0.086 | 0.762 | 0.739 | -0.775 |
| 03 | -1.038 | 1.726 | -1.966 | -2.588 | 0.483 | -0.228 | -4.371 | -3.136 | -2.546 | 1.828 | -4.371 | -3.121 | -4.371 | -0.094 | -3.042 | -4.371 |
| 04 | -1.622 | -2.983 | -3.134 | 0.573 | 2.325 | 0.279 | -0.339 | 0.254 | -3.042 | -2.225 | -4.371 | -3.111 | -4.371 | -3.121 | -2.591 | -3.129 |
| 05 | 1.495 | -2.2 | 0.813 | 1.296 | -1.453 | -3.09 | -4.371 | 0.209 | -1.596 | -4.371 | -2.579 | -0.593 | -0.998 | 0.002 | -1.074 | 0.364 |
| 06 | -0.165 | -3.097 | 1.473 | -4.371 | -4.371 | -4.371 | 0.125 | -4.371 | -4.371 | -4.371 | 0.961 | -3.094 | -3.125 | -4.371 | 1.918 | -4.371 |
| 07 | -4.371 | -4.371 | -4.371 | -0.129 | -3.097 | -4.371 | -4.371 | -4.371 | 1.327 | 1.657 | -2.594 | 1.763 | -3.094 | -4.371 | -4.371 | -4.371 |
| 08 | 0.795 | 0.215 | -3.109 | -0.985 | -0.014 | 1.197 | -3.088 | -0.065 | -4.371 | -2.594 | -4.371 | -4.371 | 0.579 | 1.07 | -1.014 | 0.514 |
| 09 | -1.085 | -1.593 | 0.018 | 1.235 | 0.683 | -1.774 | -1.31 | 1.634 | -2.219 | -4.371 | -2.582 | -1.315 | -0.788 | -3.021 | 0.326 | 0.1 |
| 10 | -2.204 | -1.223 | 0.86 | -2.181 | -4.371 | -3.097 | -1.011 | -4.371 | -1.004 | -1.098 | 0.662 | -2.204 | -0.371 | 0.365 | 2.008 | -1.023 |
| 11 | -1.971 | -1.019 | -0.406 | -4.371 | -0.142 | 0.06 | -2.237 | -2.552 | 0.647 | 0.418 | 2.021 | 0.13 | -2.199 | -4.371 | -0.848 | -3.125 |
| 12 | 0.607 | -3.112 | -0.205 | -1.095 | 1.002 | -3.123 | -3.054 | -1.984 | 1.582 | -2.217 | 1.127 | -0.378 | -1.084 | -3.113 | -0.205 | -2.579 |
| 13 | -0.334 | 2.184 | -2.992 | -2.164 | -2.579 | -1.96 | -4.371 | -4.371 | -4.371 | 1.553 | -4.371 | -4.371 | -4.371 | 0.178 | -4.371 | -4.371 |
| 14 | -2.094 | -2.575 | -4.371 | -0.502 | 1.838 | 0.572 | -1.204 | 1.885 | -4.371 | -2.992 | -4.371 | -4.371 | -2.473 | -4.371 | -4.371 | -3.142 |
| 15 | -0.836 | -0.155 | -0.208 | 1.482 | -3.093 | -1.977 | -4.371 | 0.539 | -3.108 | -4.371 | -3.108 | -1.445 | -1.093 | 0.05 | -1.64 | 1.735 |
| 16 | -4.371 | -4.371 | -0.198 | -4.371 | -2.236 | -3.021 | 0.62 | -3.135 | -4.371 | -4.371 | 0.025 | -4.371 | -1.648 | -4.371 | 2.468 | -4.371 |
| 17 | -2.515 | -2.085 | -4.371 | -2.236 | -4.371 | -3.021 | -4.371 | -4.371 | 1.214 | 1.902 | -2.227 | 1.673 | -4.371 | -3.135 | -4.371 | -4.371 |
| 18 | 0.994 | -0.924 | -1.96 | -1.448 | 0.825 | 1.055 | -1.758 | 0.465 | -3.133 | -3.122 | -3.09 | -4.371 | -0.241 | 0.71 | -0.034 | 0.516 |
| 19 | -0.638 | -0.005 | 0.591 | 0.909 | 0.097 | 0.376 | -1.455 | 0.938 | -1.977 | -1.018 | -1.098 | -0.709 | -0.585 | 0.203 | -0.064 | -0.228 |
| 20 | -0.992 | -1.084 | -0.034 | -0.432 | 0.5 | 0.028 | -1.109 | 0.03 | 0.374 | -0.476 | -0.295 | -0.707 | -0.831 | -0.156 | 1.371 | 0.09 |
| 21 | -0.916 | -0.012 | 0.132 | 0.321 | 0.131 | -0.654 | -1.743 | 0.008 | -0.123 | 0.104 | 1.156 | -0.207 | -1.087 | -0.068 | 0.114 | -0.135 |
| 22 | -0.069 | -0.708 | 0.033 | -1.195 | -0.066 | -0.176 | -1.976 | 0.493 | 1.071 | -0.16 | 0.229 | -0.533 | -0.923 | -0.25 | 0.289 | 0.447 |