Model info
| Transcription factor | Olig2 | ||||||||
| Model | OLIG2_MOUSE.H10MO.B | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 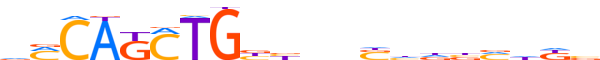 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 20 | ||||||||
| Quality | B | ||||||||
| Consensus | vMCAKCTGbbnnShdbvhdb | ||||||||
| wAUC | 0.7955124233144746 | ||||||||
| Best AUC | 0.8161175869990306 | ||||||||
| Benchmark datasets | 2 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | Tal-related factors{1.2.3} | ||||||||
| TF subfamily | Neurogenin / Atonal-like factors{1.2.3.4} | ||||||||
| MGI | 1355331 | ||||||||
| EntrezGene | 50913 | ||||||||
| UniProt ID | OLIG2_MOUSE | ||||||||
| UniProt AC | Q9EQW6 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 170.995 | 98.153 | 176.891 | 53.961 |
| 02 | 121.367 | 280.337 | 95.573 | 2.724 |
| 03 | 48.937 | 448.132 | 1.911 | 1.021 |
| 04 | 447.481 | 0.0 | 11.926 | 40.593 |
| 05 | 6.213 | 5.014 | 337.26 | 151.513 |
| 06 | 88.775 | 393.876 | 15.178 | 2.171 |
| 07 | 22.933 | 11.183 | 3.747 | 462.137 |
| 08 | 1.089 | 0.0 | 482.289 | 16.622 |
| 09 | 9.6 | 195.853 | 143.041 | 151.506 |
| 10 | 53.364 | 111.097 | 80.693 | 254.845 |
| 11 | 85.673 | 125.595 | 138.796 | 149.936 |
| 12 | 90.203 | 154.577 | 143.788 | 111.432 |
| 13 | 53.482 | 306.876 | 77.475 | 62.167 |
| 14 | 150.978 | 181.978 | 39.766 | 127.279 |
| 15 | 140.821 | 29.348 | 217.461 | 112.37 |
| 16 | 25.231 | 163.63 | 204.476 | 106.663 |
| 17 | 103.967 | 275.886 | 79.557 | 40.59 |
| 18 | 110.728 | 88.194 | 65.254 | 235.825 |
| 19 | 63.157 | 39.361 | 291.71 | 105.773 |
| 20 | 50.86 | 151.279 | 208.833 | 89.029 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 0.31 | -0.238 | 0.344 | -0.824 |
| 02 | -0.029 | 0.801 | -0.265 | -3.387 |
| 03 | -0.919 | 1.268 | -3.598 | -3.895 |
| 04 | 1.266 | -4.4 | -2.239 | -1.1 |
| 05 | -2.791 | -2.958 | 0.985 | 0.19 |
| 06 | -0.337 | 1.139 | -2.023 | -3.526 |
| 07 | -1.643 | -2.296 | -3.173 | 1.299 |
| 08 | -3.869 | -4.4 | 1.341 | -1.941 |
| 09 | -2.429 | 0.445 | 0.133 | 0.19 |
| 10 | -0.835 | -0.116 | -0.431 | 0.706 |
| 11 | -0.372 | 0.005 | 0.103 | 0.18 |
| 12 | -0.322 | 0.21 | 0.138 | -0.113 |
| 13 | -0.833 | 0.891 | -0.471 | -0.686 |
| 14 | 0.187 | 0.372 | -1.119 | 0.018 |
| 15 | 0.118 | -1.41 | 0.548 | -0.105 |
| 16 | -1.553 | 0.266 | 0.487 | -0.157 |
| 17 | -0.182 | 0.785 | -0.445 | -1.1 |
| 18 | -0.12 | -0.344 | -0.639 | 0.629 |
| 19 | -0.671 | -1.129 | 0.84 | -0.165 |
| 20 | -0.882 | 0.189 | 0.508 | -0.334 |