Model info
| Transcription factor | Nr1d1 | ||||||||
| Model | NR1D1_MOUSE.H10DI.D | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 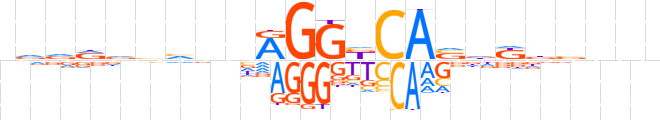 | ||||||||
| LOGO (reverse complement) | 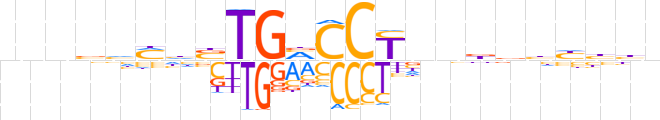 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | D | ||||||||
| Consensus | nvdddvvnnRGGbCAvdKdbnnn | ||||||||
| wAUC | 0.7268982058707454 | ||||||||
| Best AUC | 0.7268982058707455 | ||||||||
| Benchmark datasets | 1 | ||||||||
| Aligned words | 513 | ||||||||
| TF family | Thyroid hormone receptor-related factors (NR1){2.1.2} | ||||||||
| TF subfamily | Rev-ErbA (NR1D){2.1.2.3} | ||||||||
| MGI | 2444210 | ||||||||
| EntrezGene | 217166 | ||||||||
| UniProt ID | NR1D1_MOUSE | ||||||||
| UniProt AC | Q3UV55 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 42.814 | 17.61 | 55.57 | 6.77 | 42.866 | 14.594 | 7.976 | 22.326 | 56.48 | 21.262 | 47.384 | 18.046 | 38.14 | 18.449 | 72.705 | 16.004 |
| 02 | 40.802 | 22.139 | 106.191 | 11.167 | 46.386 | 10.599 | 4.325 | 10.604 | 51.322 | 18.892 | 82.895 | 30.527 | 7.149 | 2.207 | 40.767 | 13.023 |
| 03 | 31.109 | 11.248 | 93.222 | 10.081 | 28.793 | 4.239 | 5.999 | 14.806 | 49.184 | 24.855 | 134.387 | 25.752 | 1.896 | 5.827 | 43.858 | 13.74 |
| 04 | 37.281 | 4.87 | 52.784 | 16.047 | 13.406 | 16.665 | 4.883 | 11.214 | 75.094 | 13.451 | 115.045 | 73.877 | 17.596 | 13.576 | 18.794 | 14.414 |
| 05 | 54.459 | 33.837 | 34.074 | 21.007 | 22.227 | 16.605 | 0.961 | 8.768 | 46.887 | 73.453 | 39.919 | 31.247 | 15.622 | 64.683 | 23.671 | 11.575 |
| 06 | 64.889 | 31.47 | 32.085 | 10.75 | 107.928 | 59.144 | 4.895 | 16.612 | 23.329 | 41.846 | 26.189 | 7.262 | 16.599 | 27.189 | 14.287 | 14.522 |
| 07 | 43.877 | 52.392 | 90.823 | 25.653 | 77.854 | 29.034 | 10.646 | 42.115 | 27.158 | 10.875 | 24.568 | 14.854 | 12.037 | 13.414 | 19.73 | 3.966 |
| 08 | 55.406 | 29.377 | 55.535 | 20.608 | 37.677 | 24.78 | 7.787 | 35.471 | 30.009 | 41.618 | 34.224 | 39.916 | 7.964 | 20.132 | 41.173 | 17.32 |
| 09 | 97.114 | 0.0 | 33.053 | 0.889 | 104.759 | 0.0 | 5.025 | 6.122 | 97.592 | 0.0 | 32.014 | 9.113 | 59.707 | 0.0 | 48.444 | 5.164 |
| 10 | 0.0 | 1.111 | 358.062 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 117.577 | 0.959 | 0.0 | 1.113 | 19.22 | 0.955 |
| 11 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 2.223 | 0.0 | 7.544 | 5.014 | 428.928 | 53.372 | 0.0 | 0.0 | 1.914 | 0.0 |
| 12 | 0.0 | 1.816 | 2.836 | 2.892 | 0.0 | 2.046 | 0.0 | 2.968 | 41.136 | 49.774 | 91.578 | 250.578 | 0.917 | 11.59 | 6.249 | 34.616 |
| 13 | 2.783 | 38.326 | 0.0 | 0.944 | 1.002 | 63.06 | 0.0 | 1.165 | 0.0 | 98.806 | 1.858 | 0.0 | 0.961 | 278.437 | 6.02 | 5.636 |
| 14 | 4.746 | 0.0 | 0.0 | 0.0 | 460.376 | 2.83 | 2.919 | 12.504 | 6.997 | 0.0 | 0.88 | 0.0 | 6.736 | 0.0 | 0.0 | 1.009 |
| 15 | 91.279 | 122.234 | 248.759 | 16.583 | 0.912 | 0.955 | 0.964 | 0.0 | 0.864 | 0.87 | 2.065 | 0.0 | 2.999 | 6.684 | 2.808 | 1.022 |
| 16 | 36.409 | 3.085 | 37.239 | 19.32 | 42.547 | 10.845 | 7.234 | 70.118 | 62.408 | 31.955 | 100.398 | 59.836 | 2.97 | 0.0 | 8.724 | 5.911 |
| 17 | 20.639 | 8.109 | 102.063 | 13.523 | 18.299 | 7.036 | 4.728 | 15.82 | 28.723 | 7.463 | 88.452 | 28.957 | 10.075 | 6.283 | 112.549 | 26.277 |
| 18 | 13.767 | 9.397 | 49.569 | 5.003 | 8.74 | 3.012 | 5.697 | 11.442 | 65.339 | 46.037 | 115.748 | 80.669 | 9.978 | 4.631 | 51.013 | 18.955 |
| 19 | 11.289 | 28.17 | 47.188 | 11.177 | 14.186 | 19.956 | 5.009 | 23.926 | 31.826 | 50.038 | 116.981 | 23.182 | 12.539 | 44.193 | 36.043 | 23.294 |
| 20 | 10.759 | 18.765 | 31.303 | 9.014 | 55.622 | 43.893 | 11.148 | 31.694 | 40.536 | 70.13 | 57.485 | 37.07 | 9.399 | 32.931 | 20.72 | 18.529 |
| 21 | 37.753 | 29.403 | 39.099 | 10.06 | 75.757 | 39.184 | 11.142 | 39.636 | 46.251 | 30.043 | 28.319 | 16.043 | 11.201 | 18.901 | 36.825 | 29.38 |
| 22 | 45.94 | 35.654 | 65.426 | 23.942 | 53.436 | 26.715 | 4.273 | 33.106 | 30.457 | 41.428 | 31.532 | 11.969 | 12.498 | 14.991 | 49.441 | 18.19 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.314 | -0.562 | 0.572 | -1.484 | 0.315 | -0.746 | -1.328 | -0.329 | 0.588 | -0.377 | 0.414 | -0.538 | 0.199 | -0.517 | 0.839 | -0.656 |
| 02 | 0.266 | -0.338 | 1.217 | -1.005 | 0.393 | -1.056 | -1.902 | -1.055 | 0.493 | -0.493 | 0.97 | -0.021 | -1.432 | -2.499 | 0.265 | -0.856 |
| 03 | -0.002 | -0.998 | 1.087 | -1.104 | -0.079 | -1.92 | -1.598 | -0.731 | 0.451 | -0.224 | 1.451 | -0.189 | -2.626 | -1.625 | 0.337 | -0.804 |
| 04 | 0.176 | -1.793 | 0.521 | -0.653 | -0.828 | -0.616 | -1.79 | -1.001 | 0.872 | -0.825 | 1.296 | 0.855 | -0.563 | -0.816 | -0.498 | -0.758 |
| 05 | 0.552 | 0.081 | 0.087 | -0.389 | -0.334 | -0.62 | -3.152 | -1.238 | 0.404 | 0.85 | 0.244 | 0.002 | -0.679 | 0.723 | -0.272 | -0.971 |
| 06 | 0.726 | 0.009 | 0.028 | -1.042 | 1.233 | 0.634 | -1.788 | -0.619 | -0.286 | 0.291 | -0.172 | -1.418 | -0.62 | -0.135 | -0.766 | -0.75 |
| 07 | 0.338 | 0.514 | 1.061 | -0.193 | 0.907 | -0.071 | -1.051 | 0.297 | -0.137 | -1.031 | -0.235 | -0.728 | -0.933 | -0.828 | -0.451 | -1.981 |
| 08 | 0.569 | -0.059 | 0.572 | -0.408 | 0.187 | -0.227 | -1.351 | 0.127 | -0.038 | 0.285 | 0.092 | 0.244 | -1.33 | -0.431 | 0.275 | -0.578 |
| 09 | 1.127 | -4.398 | 0.057 | -3.208 | 1.203 | -4.398 | -1.764 | -1.579 | 1.132 | -4.398 | 0.026 | -1.201 | 0.644 | -4.398 | 0.436 | -1.738 |
| 10 | -4.398 | -3.048 | 2.429 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | 1.318 | -3.154 | -4.398 | -3.046 | -0.476 | -3.157 |
| 11 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -4.398 | -2.492 | -4.398 | -1.381 | -1.765 | 2.61 | 0.532 | -4.398 | -4.398 | -2.618 | -4.398 |
| 12 | -4.398 | -2.662 | -2.282 | -2.264 | -4.398 | -2.563 | -4.398 | -2.242 | 0.274 | 0.463 | 1.069 | 2.073 | -3.186 | -0.969 | -1.56 | 0.103 |
| 13 | -2.298 | 0.204 | -4.398 | -3.166 | -3.123 | 0.698 | -4.398 | -3.012 | -4.398 | 1.145 | -2.643 | -4.398 | -3.152 | 2.178 | -1.595 | -1.657 |
| 14 | -1.816 | -4.398 | -4.398 | -4.398 | 2.681 | -2.283 | -2.256 | -0.896 | -1.453 | -4.398 | -3.215 | -4.398 | -1.489 | -4.398 | -4.398 | -3.118 |
| 15 | 1.066 | 1.357 | 2.066 | -0.621 | -3.19 | -3.157 | -3.151 | -4.398 | -3.228 | -3.222 | -2.555 | -4.398 | -2.233 | -1.496 | -2.29 | -3.109 |
| 16 | 0.153 | -2.207 | 0.175 | -0.471 | 0.307 | -1.034 | -1.421 | 0.803 | 0.688 | 0.024 | 1.161 | 0.646 | -2.241 | -4.398 | -1.243 | -1.612 |
| 17 | -0.407 | -1.313 | 1.177 | -0.82 | -0.525 | -1.448 | -1.82 | -0.667 | -0.081 | -1.392 | 1.034 | -0.073 | -1.104 | -1.555 | 1.274 | -0.169 |
| 18 | -0.802 | -1.171 | 0.459 | -1.768 | -1.241 | -2.228 | -1.647 | -0.982 | 0.733 | 0.385 | 1.302 | 0.943 | -1.114 | -1.839 | 0.487 | -0.49 |
| 19 | -0.995 | -0.1 | 0.41 | -1.004 | -0.773 | -0.44 | -1.766 | -0.261 | 0.02 | 0.468 | 1.313 | -0.292 | -0.893 | 0.345 | 0.143 | -0.288 |
| 20 | -1.041 | -0.5 | 0.004 | -1.211 | 0.573 | 0.338 | -1.007 | 0.016 | 0.259 | 0.803 | 0.606 | 0.171 | -1.171 | 0.054 | -0.403 | -0.512 |
| 21 | 0.189 | -0.058 | 0.224 | -1.106 | 0.88 | 0.226 | -1.007 | 0.237 | 0.39 | -0.037 | -0.095 | -0.653 | -1.002 | -0.493 | 0.164 | -0.059 |
| 22 | 0.383 | 0.132 | 0.734 | -0.261 | 0.533 | -0.153 | -1.913 | 0.059 | -0.023 | 0.281 | 0.011 | -0.938 | -0.896 | -0.719 | 0.456 | -0.53 |