Model info
| Transcription factor | Mxi1 | ||||||||
| Model | MXI1_MOUSE.H10DI.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 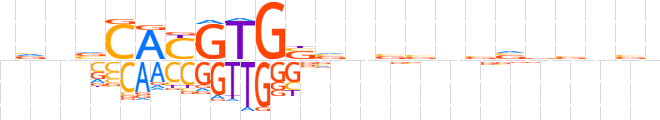 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | B | ||||||||
| Consensus | nvvvCAYGTGSvnvvnbvvvnbn | ||||||||
| wAUC | 0.7838391847178962 | ||||||||
| Best AUC | 0.8008708723194874 | ||||||||
| Benchmark datasets | 2 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | bHLH-ZIP factors{1.2.6} | ||||||||
| TF subfamily | Mad-like factors{1.2.6.7} | ||||||||
| MGI | 97245 | ||||||||
| EntrezGene | 17859 | ||||||||
| UniProt ID | MXI1_MOUSE | ||||||||
| UniProt AC | P50540 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 52.049 | 13.545 | 54.0 | 7.498 | 42.982 | 40.415 | 33.65 | 4.755 | 33.55 | 37.895 | 76.471 | 18.538 | 19.454 | 11.856 | 45.73 | 7.613 |
| 02 | 55.409 | 29.234 | 55.271 | 8.121 | 45.825 | 16.917 | 19.084 | 21.884 | 62.077 | 45.954 | 69.779 | 32.04 | 8.859 | 2.185 | 19.046 | 8.314 |
| 03 | 35.952 | 54.695 | 78.434 | 3.089 | 16.91 | 55.242 | 18.084 | 4.054 | 29.296 | 64.419 | 51.986 | 17.48 | 10.896 | 17.979 | 37.68 | 3.805 |
| 04 | 2.106 | 81.362 | 9.585 | 0.0 | 0.0 | 188.316 | 4.019 | 0.0 | 2.674 | 131.224 | 51.368 | 0.918 | 0.0 | 28.428 | 0.0 | 0.0 |
| 05 | 2.106 | 0.0 | 2.674 | 0.0 | 381.02 | 6.886 | 37.528 | 3.897 | 24.283 | 4.994 | 34.763 | 0.931 | 0.918 | 0.0 | 0.0 | 0.0 |
| 06 | 12.427 | 321.28 | 33.903 | 40.718 | 0.932 | 7.933 | 0.93 | 2.084 | 1.761 | 53.048 | 12.214 | 7.942 | 0.0 | 2.953 | 0.944 | 0.93 |
| 07 | 0.0 | 2.266 | 12.854 | 0.0 | 44.159 | 6.358 | 331.687 | 3.01 | 4.079 | 0.0 | 43.912 | 0.0 | 1.95 | 0.997 | 48.727 | 0.0 |
| 08 | 4.745 | 0.0 | 0.0 | 45.443 | 0.0 | 0.0 | 0.885 | 8.736 | 29.441 | 6.724 | 7.667 | 393.349 | 0.812 | 0.0 | 0.0 | 2.198 |
| 09 | 0.0 | 0.0 | 34.997 | 0.0 | 0.0 | 0.0 | 5.721 | 1.003 | 0.0 | 0.0 | 8.552 | 0.0 | 1.205 | 0.959 | 446.453 | 1.11 |
| 10 | 0.0 | 0.0 | 1.205 | 0.0 | 0.0 | 0.0 | 0.959 | 0.0 | 13.932 | 113.606 | 280.43 | 87.755 | 0.0 | 1.11 | 1.003 | 0.0 |
| 11 | 1.935 | 6.036 | 5.961 | 0.0 | 22.284 | 18.498 | 61.215 | 12.719 | 69.754 | 91.765 | 105.878 | 16.2 | 11.145 | 17.575 | 45.157 | 13.878 |
| 12 | 25.756 | 8.312 | 57.837 | 13.213 | 44.045 | 28.662 | 25.761 | 35.405 | 54.075 | 37.95 | 71.933 | 54.254 | 7.227 | 8.137 | 18.602 | 8.831 |
| 13 | 12.364 | 27.368 | 78.537 | 12.833 | 5.806 | 27.675 | 32.055 | 17.526 | 41.18 | 60.229 | 54.058 | 18.666 | 12.76 | 18.702 | 70.307 | 9.934 |
| 14 | 16.575 | 17.367 | 34.069 | 4.1 | 8.168 | 65.761 | 33.025 | 27.019 | 30.112 | 85.222 | 98.015 | 21.607 | 9.438 | 15.875 | 26.337 | 7.309 |
| 15 | 20.051 | 20.539 | 17.148 | 6.555 | 34.27 | 59.654 | 43.593 | 46.707 | 27.381 | 62.309 | 80.812 | 20.944 | 1.056 | 9.933 | 35.044 | 14.003 |
| 16 | 7.971 | 17.368 | 49.471 | 7.948 | 24.651 | 40.986 | 42.301 | 44.497 | 33.216 | 41.935 | 84.617 | 16.829 | 11.824 | 17.324 | 48.451 | 10.611 |
| 17 | 10.511 | 14.479 | 49.94 | 2.733 | 37.233 | 37.518 | 32.117 | 10.745 | 41.473 | 66.095 | 110.286 | 6.984 | 9.123 | 13.229 | 50.763 | 6.771 |
| 18 | 16.358 | 12.95 | 55.772 | 13.261 | 30.517 | 32.703 | 32.484 | 35.617 | 44.526 | 58.397 | 106.247 | 33.936 | 2.128 | 11.678 | 9.695 | 3.732 |
| 19 | 21.76 | 13.032 | 50.191 | 8.545 | 19.216 | 35.942 | 43.323 | 17.247 | 51.0 | 56.234 | 71.843 | 25.121 | 10.545 | 33.377 | 34.825 | 7.799 |
| 20 | 24.278 | 19.733 | 45.772 | 12.738 | 41.635 | 39.781 | 31.353 | 25.817 | 41.533 | 63.914 | 57.896 | 36.84 | 13.031 | 9.981 | 30.592 | 5.107 |
| 21 | 17.135 | 24.428 | 68.153 | 10.761 | 21.104 | 44.853 | 30.434 | 37.017 | 21.004 | 61.776 | 70.366 | 12.467 | 5.042 | 12.39 | 53.328 | 9.743 |
| 22 | 18.518 | 15.971 | 26.829 | 2.968 | 36.789 | 40.803 | 31.7 | 34.155 | 35.378 | 73.932 | 85.829 | 27.142 | 6.028 | 24.963 | 22.242 | 16.755 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.505 | -0.82 | 0.542 | -1.389 | 0.315 | 0.254 | 0.073 | -1.817 | 0.07 | 0.191 | 0.888 | -0.514 | -0.467 | -0.949 | 0.377 | -1.375 |
| 02 | 0.567 | -0.066 | 0.565 | -1.313 | 0.379 | -0.603 | -0.485 | -0.351 | 0.68 | 0.382 | 0.797 | 0.025 | -1.23 | -2.509 | -0.487 | -1.291 |
| 03 | 0.139 | 0.554 | 0.913 | -2.208 | -0.604 | 0.564 | -0.538 | -1.963 | -0.064 | 0.717 | 0.504 | -0.571 | -1.031 | -0.544 | 0.185 | -2.021 |
| 04 | -2.54 | 0.949 | -1.154 | -4.4 | -4.4 | 1.786 | -1.971 | -4.4 | -2.335 | 1.425 | 0.492 | -3.187 | -4.4 | -0.093 | -4.4 | -4.4 |
| 05 | -2.54 | -4.4 | -2.335 | -4.4 | 2.489 | -1.47 | 0.181 | -1.999 | -0.249 | -1.771 | 0.105 | -3.177 | -3.187 | -4.4 | -4.4 | -4.4 |
| 06 | -0.904 | 2.319 | 0.081 | 0.262 | -3.176 | -1.336 | -3.178 | -2.549 | -2.689 | 0.524 | -0.92 | -1.334 | -4.4 | -2.248 | -3.167 | -3.178 |
| 07 | -4.4 | -2.478 | -0.871 | -4.4 | 0.342 | -1.545 | 2.351 | -2.231 | -1.957 | -4.4 | 0.337 | -4.4 | -2.605 | -3.128 | 0.44 | -4.4 |
| 08 | -1.819 | -4.4 | -4.4 | 0.371 | -4.4 | -4.4 | -3.213 | -1.243 | -0.059 | -1.493 | -1.368 | 2.521 | -3.272 | -4.4 | -4.4 | -2.504 |
| 09 | -4.4 | -4.4 | 0.112 | -4.4 | -4.4 | -4.4 | -1.645 | -3.124 | -4.4 | -4.4 | -1.264 | -4.4 | -2.988 | -3.156 | 2.648 | -3.05 |
| 10 | -4.4 | -4.4 | -2.988 | -4.4 | -4.4 | -4.4 | -3.156 | -4.4 | -0.793 | 1.282 | 2.183 | 1.025 | -4.4 | -3.05 | -3.124 | -4.4 |
| 11 | -2.611 | -1.594 | -1.606 | -4.4 | -0.333 | -0.516 | 0.666 | -0.881 | 0.796 | 1.069 | 1.212 | -0.646 | -1.009 | -0.566 | 0.364 | -0.796 |
| 12 | -0.191 | -1.291 | 0.61 | -0.844 | 0.34 | -0.085 | -0.191 | 0.123 | 0.543 | 0.192 | 0.827 | 0.546 | -1.424 | -1.311 | -0.51 | -1.233 |
| 13 | -0.909 | -0.131 | 0.914 | -0.873 | -1.631 | -0.12 | 0.025 | -0.569 | 0.273 | 0.65 | 0.543 | -0.507 | -0.878 | -0.505 | 0.804 | -1.12 |
| 14 | -0.623 | -0.578 | 0.085 | -1.953 | -1.308 | 0.738 | 0.055 | -0.144 | -0.037 | 0.995 | 1.135 | -0.364 | -1.169 | -0.665 | -0.169 | -1.413 |
| 15 | -0.437 | -0.413 | -0.59 | -1.517 | 0.091 | 0.641 | 0.329 | 0.398 | -0.13 | 0.684 | 0.943 | -0.394 | -3.087 | -1.12 | 0.113 | -0.788 |
| 16 | -1.331 | -0.578 | 0.455 | -1.334 | -0.234 | 0.268 | 0.3 | 0.35 | 0.06 | 0.291 | 0.988 | -0.608 | -0.952 | -0.58 | 0.434 | -1.057 |
| 17 | -1.066 | -0.755 | 0.464 | -2.316 | 0.173 | 0.181 | 0.027 | -1.044 | 0.28 | 0.743 | 1.252 | -1.457 | -1.202 | -0.843 | 0.48 | -1.486 |
| 18 | -0.636 | -0.864 | 0.574 | -0.841 | -0.023 | 0.045 | 0.038 | 0.129 | 0.35 | 0.62 | 1.215 | 0.081 | -2.532 | -0.964 | -1.144 | -2.038 |
| 19 | -0.357 | -0.858 | 0.469 | -1.265 | -0.479 | 0.138 | 0.323 | -0.584 | 0.485 | 0.582 | 0.825 | -0.215 | -1.063 | 0.065 | 0.107 | -1.352 |
| 20 | -0.249 | -0.453 | 0.378 | -0.88 | 0.284 | 0.239 | 0.003 | -0.188 | 0.281 | 0.709 | 0.611 | 0.163 | -0.858 | -1.116 | -0.021 | -1.75 |
| 21 | -0.591 | -0.243 | 0.773 | -1.043 | -0.387 | 0.358 | -0.026 | 0.167 | -0.391 | 0.675 | 0.805 | -0.901 | -1.762 | -0.907 | 0.529 | -1.139 |
| 22 | -0.515 | -0.66 | -0.151 | -2.243 | 0.161 | 0.264 | 0.014 | 0.088 | 0.123 | 0.854 | 1.002 | -0.139 | -1.596 | -0.222 | -0.335 | -0.613 |