Model info
| Transcription factor | IRF4 | ||||||||
| Model | IRF4_HUMAN.H10DI.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 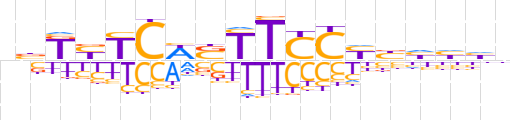 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 18 | ||||||||
| Quality | C | ||||||||
| Consensus | ndddvRGRAASWGRRMvn | ||||||||
| wAUC | 0.7537352583278552 | ||||||||
| Best AUC | 0.7663635015902608 | ||||||||
| Benchmark datasets | 2 | ||||||||
| Aligned words | 508 | ||||||||
| TF family | Interferon-regulatory factors{3.5.3} | ||||||||
| TF subfamily | IRF-4 (LSIRF, NF-EM5, MUM1, Pip){3.5.3.0.4} | ||||||||
| HGNC | 6119 | ||||||||
| EntrezGene | 3662 | ||||||||
| UniProt ID | IRF4_HUMAN | ||||||||
| UniProt AC | Q15306 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 95.055 | 6.033 | 36.364 | 21.372 | 55.377 | 9.662 | 9.676 | 19.705 | 66.336 | 17.082 | 59.288 | 20.844 | 12.864 | 3.701 | 46.529 | 20.114 |
| 02 | 132.421 | 13.324 | 73.778 | 10.109 | 13.164 | 2.942 | 8.328 | 12.044 | 57.482 | 8.534 | 78.914 | 6.927 | 9.115 | 9.045 | 33.034 | 30.841 |
| 03 | 119.405 | 5.048 | 53.882 | 33.846 | 17.317 | 5.875 | 3.542 | 7.109 | 89.761 | 11.304 | 71.967 | 21.023 | 15.991 | 8.776 | 11.401 | 23.752 |
| 04 | 50.414 | 27.895 | 149.021 | 15.143 | 17.39 | 7.639 | 4.142 | 1.832 | 22.883 | 21.418 | 70.491 | 26.002 | 19.43 | 6.932 | 55.152 | 4.216 |
| 05 | 58.134 | 5.245 | 42.683 | 4.056 | 46.028 | 2.645 | 4.898 | 10.313 | 157.392 | 12.861 | 86.181 | 22.373 | 7.374 | 2.087 | 29.631 | 8.101 |
| 06 | 31.555 | 3.149 | 230.306 | 3.917 | 17.224 | 0.0 | 5.614 | 0.0 | 32.05 | 0.0 | 128.551 | 2.791 | 6.101 | 5.145 | 32.676 | 0.92 |
| 07 | 18.345 | 1.31 | 63.926 | 3.349 | 6.423 | 0.0 | 1.871 | 0.0 | 74.313 | 13.795 | 302.516 | 6.524 | 0.0 | 0.0 | 7.628 | 0.0 |
| 08 | 87.18 | 3.166 | 8.735 | 0.0 | 14.297 | 0.0 | 0.808 | 0.0 | 346.772 | 7.954 | 16.772 | 4.444 | 3.87 | 1.295 | 3.889 | 0.818 |
| 09 | 370.278 | 0.827 | 58.729 | 22.285 | 8.65 | 2.002 | 0.9 | 0.862 | 19.626 | 1.806 | 7.955 | 0.818 | 1.672 | 0.0 | 1.696 | 1.894 |
| 10 | 58.526 | 126.465 | 208.376 | 6.858 | 0.0 | 1.166 | 3.47 | 0.0 | 5.967 | 14.03 | 47.128 | 2.155 | 2.039 | 5.892 | 16.941 | 0.987 |
| 11 | 19.678 | 4.645 | 11.566 | 30.643 | 26.562 | 11.669 | 3.645 | 105.677 | 45.569 | 11.51 | 38.7 | 180.136 | 2.108 | 0.0 | 1.771 | 6.122 |
| 12 | 16.563 | 2.992 | 73.484 | 0.878 | 9.272 | 0.0 | 16.255 | 2.297 | 5.655 | 0.862 | 49.166 | 0.0 | 5.51 | 4.288 | 308.11 | 4.67 |
| 13 | 32.819 | 0.0 | 4.18 | 0.0 | 7.323 | 0.0 | 0.0 | 0.818 | 326.377 | 8.346 | 99.806 | 12.486 | 3.027 | 0.0 | 4.817 | 0.0 |
| 14 | 204.184 | 18.769 | 140.579 | 6.014 | 1.882 | 1.688 | 3.555 | 1.222 | 21.206 | 29.779 | 55.999 | 1.819 | 0.0 | 0.818 | 10.634 | 1.852 |
| 15 | 206.821 | 3.662 | 10.589 | 6.201 | 26.177 | 10.486 | 4.69 | 9.701 | 145.495 | 33.084 | 16.264 | 15.925 | 4.982 | 3.025 | 2.038 | 0.862 |
| 16 | 55.885 | 125.324 | 181.254 | 21.012 | 19.183 | 17.874 | 3.519 | 9.68 | 5.673 | 19.777 | 5.569 | 2.562 | 2.189 | 13.491 | 12.697 | 4.311 |
| 17 | 28.3 | 10.536 | 23.166 | 20.928 | 52.924 | 50.893 | 4.618 | 68.032 | 50.326 | 45.631 | 59.017 | 48.065 | 6.028 | 9.639 | 8.311 | 13.587 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 1.104 | -1.595 | 0.15 | -0.374 | 0.567 | -1.147 | -1.145 | -0.454 | 0.746 | -0.594 | 0.635 | -0.399 | -0.87 | -2.046 | 0.394 | -0.434 |
| 02 | 1.435 | -0.836 | 0.852 | -1.103 | -0.848 | -2.251 | -1.289 | -0.934 | 0.604 | -1.266 | 0.919 | -1.464 | -1.203 | -1.21 | 0.055 | -0.013 |
| 03 | 1.331 | -1.761 | 0.54 | 0.079 | -0.58 | -1.62 | -2.086 | -1.44 | 1.047 | -0.995 | 0.827 | -0.39 | -0.658 | -1.239 | -0.987 | -0.27 |
| 04 | 0.474 | -0.112 | 1.552 | -0.712 | -0.576 | -1.372 | -1.944 | -2.657 | -0.307 | -0.372 | 0.807 | -0.181 | -0.468 | -1.464 | 0.563 | -1.927 |
| 05 | 0.615 | -1.726 | 0.308 | -1.963 | 0.383 | -2.345 | -1.789 | -1.084 | 1.607 | -0.87 | 1.007 | -0.329 | -1.405 | -2.548 | -0.053 | -1.316 |
| 06 | 0.01 | -2.191 | 1.987 | -1.994 | -0.586 | -4.4 | -1.662 | -4.4 | 0.025 | -4.4 | 1.405 | -2.298 | -1.584 | -1.744 | 0.044 | -3.186 |
| 07 | -0.524 | -2.924 | 0.709 | -2.136 | -1.536 | -4.4 | -2.639 | -4.4 | 0.859 | -0.802 | 2.259 | -1.521 | -4.4 | -4.4 | -1.373 | -4.4 |
| 08 | 1.018 | -2.186 | -1.244 | -4.4 | -0.768 | -4.4 | -3.275 | -4.4 | 2.395 | -1.333 | -0.612 | -1.879 | -2.005 | -2.933 | -2.001 | -3.267 |
| 09 | 2.461 | -3.259 | 0.625 | -0.333 | -1.253 | -2.583 | -3.201 | -3.231 | -0.458 | -2.669 | -1.333 | -3.267 | -2.732 | -4.4 | -2.72 | -2.629 |
| 10 | 0.622 | 1.389 | 1.887 | -1.474 | -4.4 | -3.013 | -2.104 | -4.4 | -1.605 | -0.786 | 0.407 | -2.521 | -2.568 | -1.617 | -0.602 | -3.136 |
| 11 | -0.455 | -1.838 | -0.973 | -0.019 | -0.16 | -0.965 | -2.06 | 1.21 | 0.373 | -0.978 | 0.211 | 1.741 | -2.54 | -4.4 | -2.684 | -1.581 |
| 12 | -0.624 | -2.237 | 0.848 | -3.218 | -1.186 | -4.4 | -0.642 | -2.467 | -1.655 | -3.231 | 0.449 | -4.4 | -1.68 | -1.912 | 2.277 | -1.833 |
| 13 | 0.048 | -4.4 | -1.935 | -4.4 | -1.412 | -4.4 | -4.4 | -3.267 | 2.335 | -1.287 | 1.153 | -0.899 | -2.226 | -4.4 | -1.805 | -4.4 |
| 14 | 1.867 | -0.502 | 1.494 | -1.598 | -2.635 | -2.724 | -2.082 | -2.978 | -0.382 | -0.048 | 0.578 | -2.662 | -4.4 | -3.267 | -1.054 | -2.648 |
| 15 | 1.879 | -2.056 | -1.059 | -1.569 | -0.175 | -1.068 | -1.829 | -1.143 | 1.528 | 0.056 | -0.642 | -0.662 | -1.773 | -2.227 | -2.568 | -3.231 |
| 16 | 0.576 | 1.38 | 1.748 | -0.391 | -0.48 | -0.55 | -2.091 | -1.145 | -1.653 | -0.45 | -1.67 | -2.372 | -2.508 | -0.824 | -0.883 | -1.907 |
| 17 | -0.098 | -1.063 | -0.295 | -0.395 | 0.522 | 0.483 | -1.844 | 0.771 | 0.472 | 0.375 | 0.63 | 0.426 | -1.595 | -1.149 | -1.291 | -0.817 |