Model info
| Transcription factor | FOXH1 | ||||||||
| Model | FOXH1_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 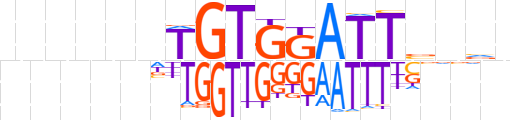 | ||||||||
| LOGO (reverse complement) | 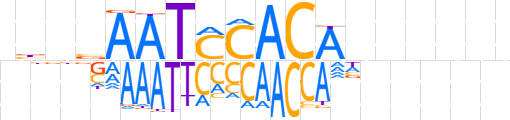 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 18 | ||||||||
| Quality | A | ||||||||
| Consensus | nnnnnnTGTGGATTbnvn | ||||||||
| wAUC | 0.9350243432791269 | ||||||||
| Best AUC | 0.9501837784769346 | ||||||||
| Benchmark datasets | 2 | ||||||||
| Aligned words | 505 | ||||||||
| TF family | Forkhead box (FOX) factors{3.3.1} | ||||||||
| TF subfamily | FOXH{3.3.1.8} | ||||||||
| HGNC | 3814 | ||||||||
| EntrezGene | 8928 | ||||||||
| UniProt ID | FOXH1_HUMAN | ||||||||
| UniProt AC | O75593 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 35.491 | 13.955 | 54.096 | 19.893 | 41.123 | 21.604 | 3.341 | 33.632 | 34.508 | 29.861 | 38.208 | 26.471 | 24.377 | 21.158 | 73.548 | 28.734 |
| 02 | 34.852 | 13.977 | 64.081 | 22.589 | 33.206 | 14.524 | 6.233 | 32.616 | 48.151 | 37.889 | 50.235 | 32.918 | 26.812 | 15.078 | 31.484 | 35.356 |
| 03 | 38.172 | 14.77 | 54.203 | 35.876 | 26.543 | 9.692 | 5.571 | 39.661 | 39.736 | 28.263 | 52.554 | 31.479 | 17.278 | 24.685 | 49.157 | 32.359 |
| 04 | 39.62 | 14.508 | 51.709 | 15.892 | 34.553 | 11.841 | 7.639 | 23.377 | 39.65 | 29.828 | 64.388 | 27.619 | 17.95 | 24.531 | 55.241 | 41.654 |
| 05 | 30.99 | 29.287 | 38.03 | 33.466 | 21.354 | 24.67 | 2.033 | 32.651 | 85.04 | 20.015 | 31.967 | 41.954 | 14.016 | 33.444 | 24.579 | 36.504 |
| 06 | 10.581 | 10.167 | 3.891 | 126.761 | 20.695 | 0.0 | 0.808 | 85.912 | 7.199 | 0.0 | 0.0 | 89.409 | 8.447 | 2.5 | 14.35 | 119.279 |
| 07 | 3.452 | 0.0 | 43.47 | 0.0 | 0.0 | 0.0 | 12.668 | 0.0 | 0.0 | 1.104 | 17.946 | 0.0 | 1.065 | 0.0 | 420.296 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 4.517 | 0.0 | 0.0 | 0.0 | 1.104 | 0.869 | 8.434 | 0.978 | 484.098 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 0.0 | 0.0 | 0.869 | 0.0 | 0.0 | 0.0 | 7.705 | 0.729 | 0.0 | 0.0 | 0.978 | 0.0 | 2.084 | 0.0 | 410.305 | 77.331 |
| 10 | 0.0 | 0.0 | 2.084 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.436 | 0.0 | 300.929 | 117.492 | 3.992 | 0.0 | 62.598 | 11.469 |
| 11 | 3.493 | 0.0 | 0.0 | 1.936 | 0.0 | 0.0 | 0.0 | 0.0 | 362.052 | 1.749 | 0.0 | 1.81 | 127.271 | 0.0 | 0.0 | 1.69 |
| 12 | 19.492 | 12.635 | 2.823 | 457.867 | 0.0 | 0.0 | 0.0 | 1.749 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.435 |
| 13 | 0.0 | 0.0 | 0.0 | 19.492 | 0.778 | 0.722 | 0.0 | 11.135 | 0.0 | 0.0 | 0.0 | 2.823 | 0.0 | 25.344 | 3.107 | 436.599 |
| 14 | 0.0 | 0.0 | 0.778 | 0.0 | 5.893 | 13.263 | 2.095 | 4.815 | 0.0 | 0.0 | 0.996 | 2.111 | 50.051 | 203.78 | 162.515 | 53.703 |
| 15 | 9.537 | 14.603 | 29.857 | 1.947 | 79.073 | 62.167 | 19.843 | 55.96 | 28.552 | 28.547 | 83.52 | 25.765 | 4.581 | 9.076 | 43.117 | 3.854 |
| 16 | 40.984 | 18.338 | 57.955 | 4.466 | 74.185 | 20.37 | 0.83 | 19.007 | 53.852 | 25.739 | 86.273 | 10.474 | 13.453 | 17.888 | 46.872 | 9.314 |
| 17 | 54.224 | 26.341 | 75.212 | 26.698 | 21.988 | 23.826 | 4.492 | 32.029 | 47.982 | 40.295 | 56.645 | 47.008 | 6.076 | 8.669 | 20.661 | 7.854 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.126 | -0.791 | 0.544 | -0.445 | 0.272 | -0.364 | -2.138 | 0.073 | 0.098 | -0.045 | 0.199 | -0.164 | -0.245 | -0.384 | 0.849 | -0.083 |
| 02 | 0.108 | -0.79 | 0.712 | -0.32 | 0.06 | -0.752 | -1.564 | 0.042 | 0.428 | 0.19 | 0.47 | 0.051 | -0.151 | -0.716 | 0.007 | 0.122 |
| 03 | 0.198 | -0.736 | 0.546 | 0.136 | -0.161 | -1.144 | -1.669 | 0.236 | 0.238 | -0.099 | 0.515 | 0.007 | -0.583 | -0.233 | 0.449 | 0.034 |
| 04 | 0.235 | -0.753 | 0.499 | -0.664 | 0.099 | -0.951 | -1.372 | -0.286 | 0.235 | -0.046 | 0.717 | -0.122 | -0.545 | -0.239 | 0.564 | 0.284 |
| 05 | -0.008 | -0.064 | 0.194 | 0.068 | -0.375 | -0.233 | -2.57 | 0.043 | 0.993 | -0.439 | 0.022 | 0.291 | -0.787 | 0.067 | -0.237 | 0.154 |
| 06 | -1.059 | -1.098 | -2.0 | 1.391 | -0.406 | -4.4 | -3.275 | 1.003 | -1.428 | -4.4 | -4.4 | 1.043 | -1.276 | -2.394 | -0.764 | 1.33 |
| 07 | -2.109 | -4.4 | 0.327 | -4.4 | -4.4 | -4.4 | -0.885 | -4.4 | -4.4 | -3.054 | -0.546 | -4.4 | -3.08 | -4.4 | 2.588 | -4.4 |
| 08 | -4.4 | -4.4 | -4.4 | -1.864 | -4.4 | -4.4 | -4.4 | -3.054 | -3.225 | -1.277 | -3.142 | 2.729 | -4.4 | -4.4 | -4.4 | -4.4 |
| 09 | -4.4 | -4.4 | -3.225 | -4.4 | -4.4 | -4.4 | -1.363 | -3.343 | -4.4 | -4.4 | -3.142 | -4.4 | -2.549 | -4.4 | 2.563 | 0.899 |
| 10 | -4.4 | -4.4 | -2.549 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -2.853 | -4.4 | 2.254 | 1.315 | -1.977 | -4.4 | 0.689 | -0.981 |
| 11 | -2.098 | -4.4 | -4.4 | -2.611 | -4.4 | -4.4 | -4.4 | -4.4 | 2.438 | -2.695 | -4.4 | -2.667 | 1.395 | -4.4 | -4.4 | -2.723 |
| 12 | -0.465 | -0.888 | -2.288 | 2.673 | -4.4 | -4.4 | -4.4 | -2.695 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -1.692 |
| 13 | -4.4 | -4.4 | -4.4 | -0.465 | -3.301 | -3.35 | -4.4 | -1.01 | -4.4 | -4.4 | -4.4 | -2.288 | -4.4 | -0.207 | -2.203 | 2.626 |
| 14 | -4.4 | -4.4 | -3.301 | -4.4 | -1.617 | -0.841 | -2.545 | -1.805 | -4.4 | -4.4 | -3.129 | -2.538 | 0.466 | 1.865 | 1.639 | 0.536 |
| 15 | -1.159 | -0.747 | -0.045 | -2.606 | 0.921 | 0.682 | -0.447 | 0.577 | -0.089 | -0.089 | 0.975 | -0.19 | -1.851 | -1.207 | 0.319 | -2.009 |
| 16 | 0.268 | -0.524 | 0.612 | -1.875 | 0.857 | -0.421 | -3.256 | -0.489 | 0.539 | -0.191 | 1.008 | -1.069 | -0.827 | -0.549 | 0.401 | -1.182 |
| 17 | 0.546 | -0.169 | 0.871 | -0.155 | -0.346 | -0.267 | -1.869 | 0.024 | 0.425 | 0.251 | 0.589 | 0.404 | -1.588 | -1.251 | -0.408 | -1.345 |