Model info
| Transcription factor | Foxa2 | ||||||||
| Model | FOXA2_MOUSE.H10DI.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 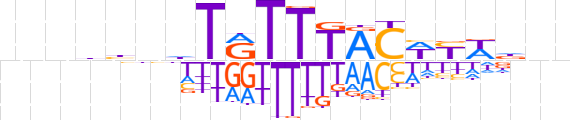 | ||||||||
| LOGO (reverse complement) | 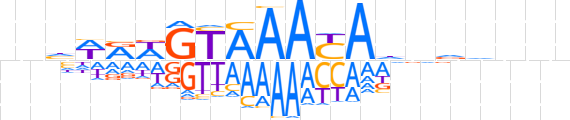 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 20 | ||||||||
| Quality | B | ||||||||
| Consensus | nnnbhnhTRTTTACWYWdnn | ||||||||
| wAUC | 0.8643172053579138 | ||||||||
| Best AUC | 0.9346536602102022 | ||||||||
| Benchmark datasets | 2 | ||||||||
| Aligned words | 511 | ||||||||
| TF family | Forkhead box (FOX) factors{3.3.1} | ||||||||
| TF subfamily | FOXA{3.3.1.1} | ||||||||
| MGI | 1347476 | ||||||||
| EntrezGene | 15376 | ||||||||
| UniProt ID | FOXA2_MOUSE | ||||||||
| UniProt AC | P35583 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 58.886 | 31.42 | 26.409 | 25.498 | 54.838 | 21.394 | 4.001 | 48.088 | 32.683 | 18.554 | 13.842 | 19.558 | 17.062 | 28.019 | 33.749 | 66.001 |
| 02 | 57.084 | 14.841 | 46.921 | 44.623 | 32.797 | 28.471 | 2.944 | 35.175 | 18.747 | 12.869 | 19.74 | 26.645 | 23.764 | 25.329 | 44.687 | 65.364 |
| 03 | 20.518 | 29.802 | 46.269 | 35.803 | 18.52 | 24.716 | 4.795 | 33.48 | 16.289 | 29.02 | 27.918 | 41.065 | 8.202 | 28.771 | 49.009 | 85.825 |
| 04 | 8.695 | 25.845 | 15.352 | 13.636 | 23.376 | 40.931 | 4.73 | 43.272 | 26.231 | 54.444 | 19.982 | 27.332 | 11.69 | 91.516 | 27.151 | 65.816 |
| 05 | 13.623 | 25.455 | 27.998 | 2.915 | 55.363 | 60.603 | 7.95 | 88.82 | 6.21 | 34.952 | 13.974 | 12.079 | 14.768 | 67.258 | 35.1 | 32.932 |
| 06 | 39.189 | 23.671 | 18.201 | 8.902 | 49.686 | 25.377 | 5.706 | 107.498 | 16.208 | 18.127 | 19.011 | 31.676 | 7.097 | 28.565 | 23.587 | 77.496 |
| 07 | 0.0 | 0.903 | 0.996 | 110.281 | 1.006 | 0.0 | 1.073 | 93.662 | 1.172 | 0.0 | 0.0 | 65.334 | 0.94 | 0.0 | 0.996 | 223.636 |
| 08 | 0.94 | 0.0 | 1.172 | 1.006 | 0.0 | 0.0 | 0.0 | 0.903 | 0.996 | 0.0 | 2.069 | 0.0 | 172.982 | 0.0 | 301.777 | 18.154 |
| 09 | 0.0 | 0.0 | 1.087 | 173.831 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 305.019 | 0.0 | 0.0 | 0.0 | 20.063 |
| 10 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.087 | 0.0 | 0.0 | 18.924 | 479.989 |
| 11 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 18.924 | 0.952 | 0.0 | 107.753 | 372.37 |
| 12 | 0.952 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 101.943 | 1.034 | 0.931 | 3.846 | 306.491 | 0.927 | 63.863 | 20.014 |
| 13 | 0.0 | 363.063 | 0.0 | 46.323 | 0.0 | 1.96 | 0.0 | 0.0 | 0.0 | 31.918 | 0.0 | 32.876 | 0.0 | 4.948 | 1.039 | 17.873 |
| 14 | 0.0 | 0.0 | 0.0 | 0.0 | 175.198 | 39.442 | 0.931 | 186.318 | 0.0 | 0.0 | 0.0 | 1.039 | 12.362 | 10.467 | 0.0 | 74.243 |
| 15 | 18.359 | 68.904 | 18.185 | 82.111 | 0.0 | 13.644 | 1.871 | 34.394 | 0.0 | 0.0 | 0.0 | 0.931 | 2.065 | 55.489 | 10.019 | 194.027 |
| 16 | 13.381 | 0.0 | 1.0 | 6.042 | 91.3 | 0.0 | 1.924 | 44.813 | 19.122 | 0.0 | 2.919 | 8.034 | 110.894 | 4.751 | 22.822 | 172.997 |
| 17 | 57.501 | 19.654 | 113.849 | 43.692 | 1.013 | 0.893 | 1.006 | 1.839 | 2.927 | 4.978 | 8.861 | 11.899 | 26.886 | 23.624 | 112.828 | 68.548 |
| 18 | 19.421 | 25.442 | 30.742 | 12.722 | 5.794 | 19.074 | 3.739 | 20.542 | 31.437 | 58.184 | 87.986 | 58.938 | 12.012 | 34.425 | 52.215 | 27.326 |
| 19 | 16.892 | 13.453 | 28.576 | 9.742 | 54.943 | 28.55 | 2.948 | 50.685 | 39.248 | 55.797 | 43.819 | 35.82 | 27.079 | 17.465 | 52.333 | 22.65 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.628 | 0.005 | -0.166 | -0.201 | 0.557 | -0.373 | -1.975 | 0.427 | 0.044 | -0.513 | -0.799 | -0.461 | -0.595 | -0.108 | 0.076 | 0.741 |
| 02 | 0.597 | -0.731 | 0.402 | 0.353 | 0.048 | -0.092 | -2.251 | 0.117 | -0.503 | -0.87 | -0.452 | -0.157 | -0.27 | -0.207 | 0.354 | 0.732 |
| 03 | -0.414 | -0.047 | 0.388 | 0.134 | -0.515 | -0.231 | -1.809 | 0.068 | -0.64 | -0.073 | -0.111 | 0.27 | -1.304 | -0.082 | 0.446 | 1.002 |
| 04 | -1.248 | -0.187 | -0.698 | -0.814 | -0.286 | 0.267 | -1.822 | 0.322 | -0.173 | 0.55 | -0.44 | -0.132 | -0.963 | 1.066 | -0.139 | 0.738 |
| 05 | -0.814 | -0.202 | -0.108 | -2.26 | 0.567 | 0.656 | -1.333 | 1.037 | -1.567 | 0.111 | -0.79 | -0.931 | -0.736 | 0.76 | 0.115 | 0.052 |
| 06 | 0.224 | -0.274 | -0.532 | -1.225 | 0.459 | -0.205 | -1.647 | 1.227 | -0.645 | -0.536 | -0.489 | 0.013 | -1.441 | -0.089 | -0.277 | 0.901 |
| 07 | -4.4 | -3.199 | -3.129 | 1.252 | -3.122 | -4.4 | -3.075 | 1.089 | -3.009 | -4.4 | -4.4 | 0.731 | -3.17 | -4.4 | -3.129 | 1.957 |
| 08 | -3.17 | -4.4 | -3.009 | -3.122 | -4.4 | -4.4 | -4.4 | -3.199 | -3.129 | -4.4 | -2.555 | -4.4 | 1.701 | -4.4 | 2.257 | -0.534 |
| 09 | -4.4 | -4.4 | -3.065 | 1.706 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | 2.267 | -4.4 | -4.4 | -4.4 | -0.436 |
| 10 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -3.065 | -4.4 | -4.4 | -0.494 | 2.72 |
| 11 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -0.494 | -3.161 | -4.4 | 1.229 | 2.467 |
| 12 | -3.161 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | 1.174 | -3.102 | -3.177 | -2.011 | 2.272 | -3.18 | 0.708 | -0.439 |
| 13 | -4.4 | 2.441 | -4.4 | 0.39 | -4.4 | -2.6 | -4.4 | -4.4 | -4.4 | 0.021 | -4.4 | 0.05 | -4.4 | -1.78 | -3.098 | -0.55 |
| 14 | -4.4 | -4.4 | -4.4 | -4.4 | 1.714 | 0.23 | -3.177 | 1.775 | -4.4 | -4.4 | -4.4 | -3.098 | -0.909 | -1.07 | -4.4 | 0.858 |
| 15 | -0.523 | 0.784 | -0.533 | 0.958 | -4.4 | -0.813 | -2.639 | 0.095 | -4.4 | -4.4 | -4.4 | -3.177 | -2.557 | 0.569 | -1.112 | 1.816 |
| 16 | -0.832 | -4.4 | -3.126 | -1.593 | 1.064 | -4.4 | -2.616 | 0.357 | -0.483 | -4.4 | -2.258 | -1.323 | 1.258 | -1.817 | -0.31 | 1.701 |
| 17 | 0.604 | -0.457 | 1.284 | 0.332 | -3.117 | -3.207 | -3.122 | -2.653 | -2.256 | -1.774 | -1.23 | -0.946 | -0.148 | -0.276 | 1.275 | 0.779 |
| 18 | -0.468 | -0.203 | -0.016 | -0.881 | -1.633 | -0.486 | -2.037 | -0.413 | 0.006 | 0.616 | 1.027 | 0.629 | -0.937 | 0.096 | 0.508 | -0.132 |
| 19 | -0.605 | -0.827 | -0.088 | -1.139 | 0.559 | -0.089 | -2.25 | 0.479 | 0.225 | 0.574 | 0.335 | 0.135 | -0.141 | -0.572 | 0.511 | -0.317 |