Model info
| Transcription factor | Evx2 | ||||||||
| Model | EVX2_MOUSE.H10DI.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 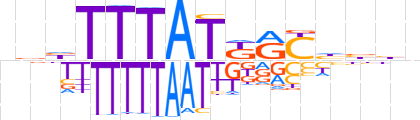 | ||||||||
| LOGO (reverse complement) | 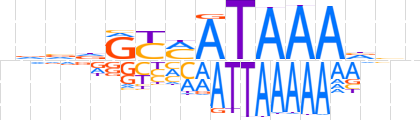 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 15 | ||||||||
| Quality | C | ||||||||
| Consensus | nbbTTTATKRYhbbn | ||||||||
| wAUC | 0.9311654294920892 | ||||||||
| Best AUC | 0.9311654294920892 | ||||||||
| Benchmark datasets | 1 | ||||||||
| Aligned words | 534 | ||||||||
| TF family | HOX-related factors{3.1.1} | ||||||||
| TF subfamily | EVX (Even-skipped homolog){3.1.1.10} | ||||||||
| MGI | 95462 | ||||||||
| EntrezGene | 14029 | ||||||||
| UniProt ID | EVX2_MOUSE | ||||||||
| UniProt AC | P49749 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 18.056 | 24.844 | 61.37 | 35.388 | 31.735 | 18.988 | 7.936 | 36.513 | 19.157 | 20.649 | 40.164 | 46.837 | 11.007 | 22.628 | 85.388 | 18.423 |
| 02 | 5.899 | 19.009 | 16.145 | 38.903 | 17.334 | 11.156 | 1.918 | 56.702 | 27.59 | 37.037 | 22.778 | 107.452 | 20.321 | 13.968 | 37.803 | 65.069 |
| 03 | 0.98 | 0.0 | 0.968 | 69.196 | 0.991 | 1.246 | 0.0 | 78.932 | 1.786 | 0.0 | 0.93 | 75.929 | 0.943 | 0.956 | 2.801 | 263.427 |
| 04 | 0.901 | 0.0 | 0.0 | 3.799 | 0.0 | 0.0 | 0.0 | 2.202 | 0.93 | 0.0 | 0.0 | 3.769 | 9.722 | 1.086 | 0.0 | 476.676 |
| 05 | 0.0 | 0.0 | 0.0 | 11.552 | 0.0 | 0.0 | 0.0 | 1.086 | 0.0 | 0.0 | 0.0 | 0.0 | 9.716 | 2.92 | 4.133 | 469.678 |
| 06 | 9.716 | 0.0 | 0.0 | 0.0 | 2.92 | 0.0 | 0.0 | 0.0 | 4.133 | 0.0 | 0.0 | 0.0 | 480.313 | 2.003 | 0.0 | 0.0 |
| 07 | 0.93 | 52.298 | 0.0 | 443.853 | 0.0 | 1.06 | 0.0 | 0.943 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 08 | 0.93 | 0.0 | 0.0 | 0.0 | 26.334 | 0.0 | 7.869 | 19.156 | 0.0 | 0.0 | 0.0 | 0.0 | 19.009 | 0.991 | 251.284 | 173.512 |
| 09 | 16.979 | 0.0 | 29.293 | 0.0 | 0.991 | 0.0 | 0.0 | 0.0 | 143.49 | 0.0 | 114.72 | 0.943 | 48.732 | 0.0 | 140.119 | 3.816 |
| 10 | 0.885 | 158.935 | 24.016 | 26.356 | 0.0 | 0.0 | 0.0 | 0.0 | 5.496 | 222.501 | 19.759 | 36.376 | 0.0 | 2.873 | 1.886 | 0.0 |
| 11 | 0.0 | 1.786 | 2.667 | 1.928 | 77.07 | 170.319 | 9.043 | 127.878 | 8.276 | 13.71 | 9.874 | 13.801 | 14.292 | 18.351 | 12.055 | 18.035 |
| 12 | 1.992 | 19.375 | 38.921 | 39.35 | 27.827 | 65.863 | 11.918 | 98.557 | 2.179 | 14.666 | 9.739 | 7.056 | 7.809 | 52.17 | 50.875 | 50.788 |
| 13 | 3.933 | 11.746 | 4.609 | 19.519 | 37.847 | 39.618 | 1.898 | 72.712 | 8.192 | 30.641 | 35.949 | 36.671 | 12.789 | 43.685 | 45.668 | 93.608 |
| 14 | 12.817 | 12.158 | 20.474 | 17.312 | 39.998 | 28.75 | 4.723 | 52.218 | 20.237 | 14.169 | 23.986 | 29.732 | 31.557 | 49.983 | 76.877 | 64.094 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.538 | -0.224 | 0.671 | 0.125 | 0.017 | -0.488 | -1.333 | 0.156 | -0.48 | -0.406 | 0.25 | 0.402 | -1.019 | -0.316 | 0.999 | -0.518 |
| 02 | -1.614 | -0.487 | -0.647 | 0.218 | -0.578 | -1.006 | -2.617 | 0.592 | -0.121 | 0.17 | -0.31 | 1.228 | -0.422 | -0.788 | 0.19 | 0.729 |
| 03 | -3.139 | -4.399 | -3.148 | 0.79 | -3.131 | -2.961 | -4.399 | 0.921 | -2.676 | -4.399 | -3.177 | 0.882 | -3.166 | -3.157 | -2.293 | 2.123 |
| 04 | -3.199 | -4.399 | -4.399 | -2.02 | -4.399 | -4.399 | -4.399 | -2.501 | -3.177 | -4.399 | -4.399 | -2.028 | -1.139 | -3.064 | -4.399 | 2.715 |
| 05 | -4.399 | -4.399 | -4.399 | -0.973 | -4.399 | -4.399 | -4.399 | -3.064 | -4.399 | -4.399 | -4.399 | -4.399 | -1.14 | -2.256 | -1.944 | 2.7 |
| 06 | -1.14 | -4.399 | -4.399 | -4.399 | -2.256 | -4.399 | -4.399 | -4.399 | -1.944 | -4.399 | -4.399 | -4.399 | 2.723 | -2.581 | -4.399 | -4.399 |
| 07 | -3.177 | 0.512 | -4.399 | 2.644 | -4.399 | -3.082 | -4.399 | -3.166 | -4.399 | -4.399 | -4.399 | -4.399 | -4.399 | -4.399 | -4.399 | -4.399 |
| 08 | -3.177 | -4.399 | -4.399 | -4.399 | -0.167 | -4.399 | -1.341 | -0.48 | -4.399 | -4.399 | -4.399 | -4.399 | -0.487 | -3.131 | 2.076 | 1.706 |
| 09 | -0.598 | -4.399 | -0.062 | -4.399 | -3.131 | -4.399 | -4.399 | -4.399 | 1.516 | -4.399 | 1.293 | -3.166 | 0.442 | -4.399 | 1.493 | -2.016 |
| 10 | -3.211 | 1.618 | -0.258 | -0.166 | -4.399 | -4.399 | -4.399 | -4.399 | -1.68 | 1.954 | -0.449 | 0.152 | -4.399 | -2.27 | -2.631 | -4.399 |
| 11 | -4.399 | -2.676 | -2.336 | -2.612 | 0.897 | 1.687 | -1.209 | 1.402 | -1.293 | -0.806 | -1.124 | -0.8 | -0.766 | -0.522 | -0.931 | -0.539 |
| 12 | -2.585 | -0.469 | 0.219 | 0.23 | -0.113 | 0.741 | -0.942 | 1.142 | -2.51 | -0.741 | -1.137 | -1.445 | -1.349 | 0.509 | 0.484 | 0.483 |
| 13 | -1.989 | -0.957 | -1.844 | -0.461 | 0.191 | 0.236 | -2.626 | 0.839 | -1.303 | -0.018 | 0.14 | 0.16 | -0.874 | 0.333 | 0.377 | 1.091 |
| 14 | -0.872 | -0.923 | -0.415 | -0.579 | 0.246 | -0.08 | -1.821 | 0.51 | -0.426 | -0.774 | -0.259 | -0.047 | 0.011 | 0.467 | 0.895 | 0.714 |