Model info
| Transcription factor | EOMES | ||||||||
| Model | EOMES_HUMAN.H10DI.D | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 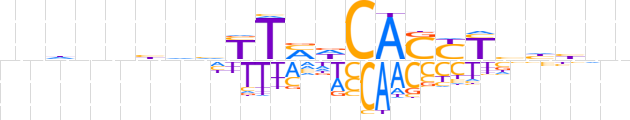 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 22 | ||||||||
| Quality | D | ||||||||
| Consensus | nnvdbRRSTGhKARdddnnhnn | ||||||||
| wAUC | 0.7822928090176371 | ||||||||
| Best AUC | 0.782292809017637 | ||||||||
| Benchmark datasets | 1 | ||||||||
| Aligned words | 376 | ||||||||
| TF family | TBrain-related factors{6.5.2} | ||||||||
| TF subfamily | TBR-2 (EOMES){6.5.2.0.2} | ||||||||
| HGNC | 3372 | ||||||||
| EntrezGene | 8320 | ||||||||
| UniProt ID | EOMES_HUMAN | ||||||||
| UniProt AC | O95936 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 17.911 | 20.026 | 45.922 | 18.01 | 23.167 | 15.971 | 2.021 | 27.932 | 26.113 | 4.037 | 31.963 | 31.96 | 22.982 | 13.779 | 41.925 | 31.279 |
| 02 | 41.817 | 9.133 | 35.209 | 4.016 | 28.727 | 6.068 | 2.153 | 16.866 | 48.29 | 30.962 | 36.52 | 6.059 | 35.3 | 6.903 | 58.985 | 7.992 |
| 03 | 24.976 | 15.928 | 83.116 | 30.114 | 21.91 | 3.072 | 3.072 | 25.012 | 32.957 | 7.149 | 68.852 | 23.909 | 6.889 | 4.937 | 18.298 | 4.81 |
| 04 | 8.993 | 13.799 | 40.951 | 22.988 | 2.986 | 16.884 | 5.134 | 6.081 | 38.228 | 36.081 | 77.136 | 21.893 | 14.909 | 5.996 | 46.105 | 16.835 |
| 05 | 54.136 | 0.0 | 6.943 | 4.036 | 58.838 | 0.0 | 1.033 | 12.89 | 122.501 | 0.0 | 25.782 | 21.044 | 45.808 | 0.0 | 14.928 | 7.062 |
| 06 | 63.669 | 4.096 | 191.594 | 21.923 | 0.0 | 0.0 | 0.0 | 0.0 | 15.984 | 1.935 | 29.827 | 0.94 | 8.056 | 3.907 | 31.101 | 1.968 |
| 07 | 1.988 | 2.966 | 80.795 | 1.961 | 1.967 | 5.93 | 1.074 | 0.967 | 8.202 | 54.74 | 188.61 | 0.969 | 1.089 | 2.025 | 16.806 | 4.911 |
| 08 | 0.0 | 0.0 | 0.0 | 13.247 | 3.919 | 0.0 | 0.0 | 61.743 | 27.66 | 0.0 | 1.947 | 257.678 | 0.0 | 0.0 | 0.0 | 8.807 |
| 09 | 0.0 | 0.0 | 31.579 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.947 | 0.0 | 0.0 | 0.958 | 340.517 | 0.0 |
| 10 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.958 | 0.0 | 0.0 | 166.547 | 56.88 | 9.956 | 140.659 | 0.0 | 0.0 | 0.0 | 0.0 |
| 11 | 3.923 | 18.671 | 48.945 | 95.008 | 3.977 | 8.983 | 0.0 | 44.878 | 0.0 | 0.0 | 7.963 | 1.994 | 1.003 | 17.021 | 54.1 | 68.535 |
| 12 | 8.903 | 0.0 | 0.0 | 0.0 | 39.678 | 1.022 | 0.0 | 3.976 | 106.718 | 0.0 | 4.29 | 0.0 | 188.435 | 0.967 | 9.993 | 11.019 |
| 13 | 261.646 | 19.122 | 42.959 | 20.006 | 1.022 | 0.967 | 0.0 | 0.0 | 8.305 | 1.988 | 2.996 | 0.993 | 11.992 | 0.0 | 3.003 | 0.0 |
| 14 | 65.108 | 24.672 | 92.918 | 100.267 | 9.116 | 4.92 | 0.0 | 8.041 | 15.927 | 7.003 | 17.994 | 8.034 | 5.066 | 1.963 | 6.966 | 7.004 |
| 15 | 34.955 | 7.04 | 24.428 | 28.795 | 25.707 | 5.957 | 0.97 | 5.924 | 43.652 | 11.902 | 29.152 | 33.171 | 10.063 | 12.29 | 49.894 | 51.1 |
| 16 | 39.767 | 11.855 | 40.724 | 22.031 | 26.121 | 7.05 | 0.988 | 3.029 | 51.929 | 7.777 | 36.817 | 7.92 | 31.328 | 23.006 | 47.715 | 16.941 |
| 17 | 42.017 | 15.956 | 65.135 | 26.038 | 17.692 | 15.992 | 0.0 | 16.005 | 48.349 | 25.816 | 41.128 | 10.951 | 14.684 | 17.852 | 12.4 | 4.985 |
| 18 | 50.947 | 20.049 | 31.986 | 19.759 | 25.83 | 23.755 | 0.0 | 26.031 | 39.017 | 29.03 | 35.376 | 15.24 | 8.1 | 12.956 | 14.901 | 22.022 |
| 19 | 43.657 | 12.074 | 26.971 | 41.193 | 23.913 | 20.225 | 0.0 | 41.652 | 33.326 | 10.981 | 5.915 | 32.041 | 15.087 | 13.111 | 19.756 | 35.097 |
| 20 | 39.581 | 12.997 | 38.572 | 24.834 | 18.162 | 16.055 | 1.018 | 21.156 | 15.846 | 6.867 | 21.913 | 8.016 | 28.774 | 23.849 | 64.362 | 32.998 |
| 21 | 30.387 | 4.938 | 40.327 | 26.712 | 24.858 | 12.961 | 1.935 | 20.014 | 33.082 | 39.071 | 33.791 | 19.922 | 15.982 | 31.991 | 27.176 | 11.854 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.264 | -0.155 | 0.665 | -0.259 | -0.011 | -0.376 | -2.298 | 0.173 | 0.107 | -1.687 | 0.306 | 0.306 | -0.019 | -0.52 | 0.575 | 0.285 |
| 02 | 0.572 | -0.918 | 0.402 | -1.692 | 0.201 | -1.308 | -2.245 | -0.323 | 0.715 | 0.275 | 0.438 | -1.309 | 0.404 | -1.186 | 0.914 | -1.046 |
| 03 | 0.063 | -0.379 | 1.255 | 0.247 | -0.066 | -1.934 | -1.934 | 0.064 | 0.336 | -1.153 | 1.067 | 0.02 | -1.188 | -1.501 | -0.243 | -1.525 |
| 04 | -0.933 | -0.519 | 0.551 | -0.019 | -1.959 | -0.322 | -1.464 | -1.306 | 0.483 | 0.426 | 1.18 | -0.067 | -0.444 | -1.319 | 0.669 | -0.325 |
| 05 | 0.828 | -4.163 | -1.18 | -1.687 | 0.911 | -4.163 | -2.831 | -0.585 | 1.641 | -4.163 | 0.094 | -0.106 | 0.663 | -4.163 | -0.442 | -1.164 |
| 06 | 0.989 | -1.673 | 2.087 | -0.066 | -4.163 | -4.163 | -4.163 | -4.163 | -0.376 | -2.335 | 0.238 | -2.9 | -1.039 | -1.717 | 0.279 | -2.32 |
| 07 | -2.312 | -1.965 | 1.226 | -2.324 | -2.321 | -1.329 | -2.802 | -2.88 | -1.021 | 0.839 | 2.072 | -2.878 | -2.792 | -2.296 | -0.327 | -1.506 |
| 08 | -4.163 | -4.163 | -4.163 | -0.559 | -1.714 | -4.163 | -4.163 | 0.959 | 0.163 | -4.163 | -2.33 | 2.383 | -4.163 | -4.163 | -4.163 | -0.953 |
| 09 | -4.163 | -4.163 | 0.294 | -4.163 | -4.163 | -4.163 | -4.163 | -4.163 | -4.163 | -4.163 | -2.33 | -4.163 | -4.163 | -2.886 | 2.662 | -4.163 |
| 10 | -4.163 | -4.163 | -4.163 | -4.163 | -4.163 | -2.886 | -4.163 | -4.163 | 1.947 | 0.877 | -0.835 | 1.779 | -4.163 | -4.163 | -4.163 | -4.163 |
| 11 | -1.713 | -0.223 | 0.728 | 1.388 | -1.7 | -0.934 | -4.163 | 0.642 | -4.163 | -4.163 | -1.05 | -2.31 | -2.852 | -0.314 | 0.828 | 1.063 |
| 12 | -0.943 | -4.163 | -4.163 | -4.163 | 0.52 | -2.839 | -4.163 | -1.701 | 1.504 | -4.163 | -1.631 | -4.163 | 2.071 | -2.879 | -0.832 | -0.737 |
| 13 | 2.398 | -0.2 | 0.599 | -0.156 | -2.839 | -2.879 | -4.163 | -4.163 | -1.01 | -2.312 | -1.956 | -2.86 | -0.655 | -4.163 | -1.954 | -4.163 |
| 14 | 1.012 | 0.051 | 1.366 | 1.442 | -0.92 | -1.504 | -4.163 | -1.04 | -0.379 | -1.172 | -0.26 | -1.041 | -1.477 | -2.323 | -1.177 | -1.172 |
| 15 | 0.395 | -1.167 | 0.041 | 0.203 | 0.091 | -1.325 | -2.877 | -1.33 | 0.615 | -0.663 | 0.215 | 0.343 | -0.825 | -0.632 | 0.747 | 0.771 |
| 16 | 0.522 | -0.666 | 0.546 | -0.061 | 0.107 | -1.166 | -2.864 | -1.946 | 0.787 | -1.072 | 0.446 | -1.055 | 0.286 | -0.018 | 0.703 | -0.319 |
| 17 | 0.577 | -0.377 | 1.012 | 0.104 | -0.276 | -0.375 | -4.163 | -0.374 | 0.716 | 0.095 | 0.556 | -0.743 | -0.458 | -0.267 | -0.623 | -1.492 |
| 18 | 0.768 | -0.154 | 0.307 | -0.168 | 0.096 | 0.013 | -4.163 | 0.103 | 0.503 | 0.211 | 0.406 | -0.422 | -1.033 | -0.58 | -0.444 | -0.061 |
| 19 | 0.615 | -0.649 | 0.138 | 0.557 | 0.02 | -0.145 | -4.163 | 0.568 | 0.347 | -0.741 | -1.332 | 0.308 | -0.432 | -0.569 | -0.168 | 0.399 |
| 20 | 0.518 | -0.577 | 0.492 | 0.057 | -0.25 | -0.371 | -2.842 | -0.101 | -0.384 | -1.191 | -0.066 | -1.043 | 0.202 | 0.017 | 1.0 | 0.338 |
| 21 | 0.256 | -1.501 | 0.536 | 0.129 | 0.058 | -0.58 | -2.335 | -0.155 | 0.34 | 0.505 | 0.361 | -0.16 | -0.376 | 0.307 | 0.146 | -0.667 |