Model info
| Transcription factor | ELK4 | ||||||||
| Model | ELK4_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 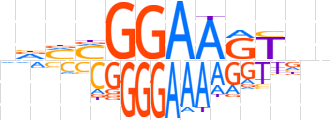 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | HT-SELEX | ||||||||
| Model length | 12 | ||||||||
| Quality | A | ||||||||
| Consensus | ndMMGGAARYvn | ||||||||
| wAUC | 0.8149096513366921 | ||||||||
| Best AUC | 0.9257179199904464 | ||||||||
| Benchmark datasets | 4 | ||||||||
| Aligned words | 11025 | ||||||||
| TF family | Ets-related factors{3.5.2} | ||||||||
| TF subfamily | Elk-like factors{3.5.2.2} | ||||||||
| HGNC | 3326 | ||||||||
| EntrezGene | 2005 | ||||||||
| UniProt ID | ELK4_HUMAN | ||||||||
| UniProt AC | P28324 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 2288.438 | 324.188 | 492.438 | 374.938 | 1171.438 | 309.188 | 448.438 | 383.938 | 1530.438 | 344.188 | 629.438 | 422.938 | 1059.438 | 310.188 | 506.438 | 427.938 |
| 02 | 586.25 | 4644.0 | 495.75 | 323.75 | 279.25 | 483.0 | 264.75 | 260.75 | 344.25 | 1069.0 | 374.75 | 288.75 | 286.25 | 747.0 | 313.75 | 262.75 |
| 03 | 285.063 | 716.813 | 250.813 | 243.313 | 552.063 | 5853.813 | 273.813 | 263.313 | 270.063 | 702.813 | 244.813 | 231.313 | 245.063 | 415.813 | 235.813 | 239.313 |
| 04 | 7.0 | 9.5 | 1324.5 | 11.25 | 19.0 | 20.5 | 7636.5 | 13.25 | 7.0 | 0.5 | 993.5 | 4.25 | 4.0 | 2.5 | 963.5 | 7.25 |
| 05 | 0.0 | 1.0 | 34.0 | 2.0 | 2.0 | 1.0 | 29.0 | 1.0 | 28.0 | 18.0 | 10723.0 | 149.0 | 1.0 | 2.0 | 32.0 | 1.0 |
| 06 | 29.0 | 1.0 | 0.0 | 1.0 | 22.0 | 0.0 | 0.0 | 0.0 | 10770.75 | 27.75 | 5.75 | 13.75 | 151.0 | 0.0 | 1.0 | 1.0 |
| 07 | 9787.188 | 62.188 | 21.188 | 1102.188 | 26.188 | 1.188 | 0.188 | 1.188 | 6.188 | 0.188 | 0.188 | 0.188 | 10.188 | 0.188 | 0.188 | 5.188 |
| 08 | 2729.438 | 185.438 | 6846.438 | 68.438 | 14.188 | 3.188 | 43.188 | 3.188 | 4.188 | 3.188 | 14.188 | 0.188 | 149.438 | 21.438 | 921.438 | 16.438 |
| 09 | 50.063 | 892.063 | 89.063 | 1866.063 | 22.813 | 47.813 | 14.813 | 127.813 | 145.813 | 1288.813 | 181.813 | 6208.813 | 10.563 | 14.563 | 11.563 | 51.563 |
| 10 | 52.063 | 51.063 | 86.063 | 40.063 | 590.313 | 337.313 | 919.313 | 396.313 | 96.063 | 63.063 | 83.063 | 55.063 | 1817.813 | 1613.813 | 3303.813 | 1518.813 |
| 11 | 731.813 | 634.813 | 811.813 | 377.813 | 618.813 | 643.813 | 516.813 | 285.813 | 1170.313 | 1225.313 | 1205.313 | 791.313 | 592.063 | 426.063 | 700.063 | 292.063 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 1.2 | -0.753 | -0.336 | -0.608 | 0.53 | -0.8 | -0.429 | -0.584 | 0.798 | -0.693 | -0.09 | -0.487 | 0.43 | -0.797 | -0.308 | -0.476 |
| 02 | -0.161 | 1.907 | -0.329 | -0.754 | -0.902 | -0.355 | -0.955 | -0.97 | -0.693 | 0.439 | -0.608 | -0.869 | -0.877 | 0.081 | -0.786 | -0.963 |
| 03 | -0.881 | 0.04 | -1.009 | -1.039 | -0.221 | 2.139 | -0.922 | -0.961 | -0.935 | 0.02 | -1.033 | -1.09 | -1.032 | -0.504 | -1.071 | -1.056 |
| 04 | -4.51 | -4.225 | 0.653 | -4.065 | -3.561 | -3.488 | 2.405 | -3.909 | -4.51 | -6.458 | 0.366 | -4.961 | -5.014 | -5.411 | 0.335 | -4.478 |
| 05 | -7.078 | -6.078 | -2.993 | -5.588 | -5.588 | -6.078 | -3.149 | -6.078 | -3.183 | -3.614 | 2.744 | -1.528 | -6.078 | -5.588 | -3.052 | -6.078 |
| 06 | -3.149 | -6.078 | -7.078 | -6.078 | -3.419 | -7.078 | -7.078 | -7.078 | 2.749 | -3.192 | -4.691 | -3.874 | -1.515 | -7.078 | -6.078 | -6.078 |
| 07 | 2.653 | -2.397 | -3.456 | 0.469 | -3.249 | -5.966 | -6.798 | -5.966 | -4.624 | -6.798 | -6.798 | -6.798 | -4.159 | -6.798 | -6.798 | -4.784 |
| 08 | 1.376 | -1.31 | 2.295 | -2.302 | -3.844 | -5.209 | -2.757 | -5.209 | -4.974 | -5.209 | -3.844 | -6.798 | -1.525 | -3.444 | 0.29 | -3.702 |
| 09 | -2.611 | 0.258 | -2.04 | 0.996 | -3.384 | -2.657 | -3.802 | -1.681 | -1.55 | 0.626 | -1.33 | 2.198 | -4.125 | -3.818 | -4.039 | -2.582 |
| 10 | -2.573 | -2.592 | -2.074 | -2.831 | -0.154 | -0.713 | 0.288 | -0.552 | -1.965 | -2.383 | -2.11 | -2.517 | 0.97 | 0.851 | 1.567 | 0.79 |
| 11 | 0.06 | -0.082 | 0.164 | -0.6 | -0.107 | -0.068 | -0.287 | -0.879 | 0.529 | 0.575 | 0.559 | 0.138 | -0.151 | -0.48 | 0.016 | -0.857 |