Model info
| Transcription factor | ELF2 | ||||||||
| Model | ELF2_HUMAN.H10DI.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 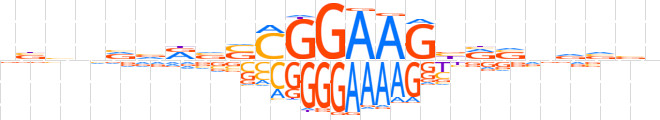 | ||||||||
| LOGO (reverse complement) | 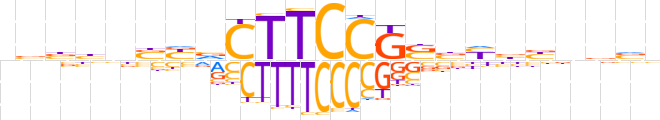 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | C | ||||||||
| Consensus | nbvnvvRvSCGGAAGbRSvvvbn | ||||||||
| wAUC | 0.8308258925973083 | ||||||||
| Best AUC | 0.8308258925973083 | ||||||||
| Benchmark datasets | 1 | ||||||||
| Aligned words | 181 | ||||||||
| TF family | Ets-related factors{3.5.2} | ||||||||
| TF subfamily | Elf-1-like factors{3.5.2.3} | ||||||||
| HGNC | 3317 | ||||||||
| EntrezGene | 1998 | ||||||||
| UniProt ID | ELF2_HUMAN | ||||||||
| UniProt AC | Q15723 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 7.979 | 8.036 | 15.692 | 1.932 | 0.0 | 17.883 | 26.139 | 9.699 | 6.146 | 11.315 | 37.162 | 8.903 | 4.092 | 8.868 | 13.067 | 4.087 |
| 02 | 10.332 | 2.919 | 4.028 | 0.938 | 8.795 | 21.282 | 8.804 | 7.221 | 25.982 | 36.738 | 21.261 | 8.079 | 5.797 | 7.2 | 3.795 | 7.83 |
| 03 | 18.733 | 17.111 | 9.124 | 5.937 | 22.083 | 19.004 | 19.19 | 7.861 | 18.21 | 7.757 | 8.965 | 2.955 | 2.802 | 7.914 | 4.995 | 8.357 |
| 04 | 24.862 | 8.061 | 23.926 | 4.978 | 5.897 | 15.996 | 27.985 | 1.909 | 7.099 | 9.165 | 24.067 | 1.944 | 3.034 | 3.954 | 17.208 | 0.915 |
| 05 | 20.741 | 5.156 | 14.006 | 0.988 | 7.979 | 13.817 | 11.977 | 3.403 | 38.457 | 13.039 | 31.874 | 9.815 | 0.914 | 0.0 | 7.899 | 0.933 |
| 06 | 42.272 | 2.867 | 15.215 | 7.737 | 10.121 | 0.955 | 16.103 | 4.833 | 35.719 | 2.947 | 20.122 | 6.968 | 4.097 | 0.0 | 7.179 | 3.865 |
| 07 | 20.845 | 27.486 | 38.847 | 5.032 | 0.998 | 2.897 | 1.915 | 0.959 | 9.702 | 11.837 | 34.999 | 2.08 | 1.888 | 7.909 | 8.785 | 4.821 |
| 08 | 0.0 | 11.678 | 17.801 | 3.954 | 5.895 | 36.317 | 5.951 | 1.965 | 5.908 | 23.132 | 54.438 | 1.068 | 1.919 | 6.944 | 4.028 | 0.0 |
| 09 | 4.88 | 8.842 | 0.0 | 0.0 | 7.906 | 70.165 | 0.0 | 0.0 | 25.618 | 54.722 | 1.879 | 0.0 | 1.829 | 4.165 | 0.994 | 0.0 |
| 10 | 0.0 | 0.0 | 39.318 | 0.914 | 4.816 | 2.048 | 124.368 | 6.662 | 1.888 | 0.0 | 0.984 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 11 | 0.0 | 0.0 | 6.705 | 0.0 | 0.0 | 0.0 | 2.048 | 0.0 | 0.0 | 0.0 | 163.719 | 0.953 | 0.0 | 1.907 | 5.669 | 0.0 |
| 12 | 0.0 | 0.0 | 0.0 | 0.0 | 1.907 | 0.0 | 0.0 | 0.0 | 168.584 | 0.0 | 9.556 | 0.0 | 0.0 | 0.0 | 0.953 | 0.0 |
| 13 | 159.644 | 0.947 | 6.935 | 2.965 | 0.0 | 0.0 | 0.0 | 0.0 | 7.667 | 0.0 | 2.841 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 14 | 19.625 | 1.928 | 145.758 | 0.0 | 0.0 | 0.0 | 0.947 | 0.0 | 3.048 | 0.991 | 5.737 | 0.0 | 0.0 | 0.0 | 2.965 | 0.0 |
| 15 | 3.838 | 13.095 | 4.785 | 0.955 | 0.0 | 1.928 | 0.0 | 0.991 | 9.665 | 51.613 | 20.962 | 73.167 | 0.0 | 0.0 | 0.0 | 0.0 |
| 16 | 5.892 | 0.961 | 4.825 | 1.825 | 13.716 | 10.936 | 33.105 | 8.88 | 5.047 | 1.973 | 15.726 | 3.0 | 13.706 | 8.123 | 52.11 | 1.174 |
| 17 | 11.29 | 3.281 | 20.853 | 2.937 | 1.011 | 6.958 | 11.964 | 2.06 | 16.045 | 19.057 | 63.511 | 7.153 | 0.9 | 4.105 | 9.874 | 0.0 |
| 18 | 9.948 | 10.286 | 7.963 | 1.048 | 2.06 | 13.2 | 9.995 | 8.147 | 39.818 | 25.766 | 30.924 | 9.696 | 0.0 | 2.851 | 9.299 | 0.0 |
| 19 | 25.875 | 6.927 | 16.109 | 2.915 | 4.962 | 11.163 | 27.056 | 8.922 | 19.883 | 6.924 | 28.429 | 2.945 | 6.221 | 1.055 | 8.744 | 2.871 |
| 20 | 16.839 | 5.918 | 32.252 | 1.931 | 6.002 | 8.003 | 10.996 | 1.068 | 13.28 | 31.209 | 28.046 | 7.802 | 0.962 | 1.926 | 12.749 | 2.016 |
| 21 | 8.037 | 8.924 | 16.065 | 4.057 | 3.81 | 18.2 | 17.01 | 8.037 | 11.833 | 27.031 | 29.146 | 16.032 | 1.043 | 6.004 | 5.77 | 0.0 |
| 22 | 3.74 | 4.142 | 13.893 | 2.949 | 17.177 | 12.068 | 13.907 | 17.008 | 10.84 | 17.711 | 29.176 | 10.265 | 4.374 | 9.836 | 8.053 | 5.862 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.337 | -0.331 | 0.319 | -1.64 | -3.578 | 0.448 | 0.822 | -0.149 | -0.587 | 0.0 | 1.17 | -0.232 | -0.969 | -0.236 | 0.14 | -0.97 |
| 02 | -0.088 | -1.277 | -0.983 | -2.221 | -0.244 | 0.619 | -0.243 | -0.433 | 0.816 | 1.158 | 0.618 | -0.326 | -0.642 | -0.436 | -1.038 | -0.356 |
| 03 | 0.493 | 0.404 | -0.208 | -0.62 | 0.655 | 0.507 | 0.517 | -0.352 | 0.465 | -0.365 | -0.225 | -1.266 | -1.314 | -0.345 | -0.783 | -0.293 |
| 04 | 0.772 | -0.328 | 0.734 | -0.786 | -0.626 | 0.338 | 0.889 | -1.65 | -0.45 | -0.204 | 0.74 | -1.635 | -1.243 | -1.001 | 0.41 | -2.239 |
| 05 | 0.593 | -0.753 | 0.208 | -2.182 | -0.337 | 0.195 | 0.055 | -1.138 | 1.204 | 0.138 | 1.018 | -0.138 | -2.24 | -3.578 | -0.347 | -2.225 |
| 06 | 1.298 | -1.294 | 0.289 | -0.367 | -0.108 | -2.208 | 0.345 | -0.814 | 1.131 | -1.269 | 0.564 | -0.467 | -0.968 | -3.578 | -0.439 | -1.022 |
| 07 | 0.598 | 0.871 | 1.214 | -0.776 | -2.174 | -1.284 | -1.648 | -2.204 | -0.149 | 0.044 | 1.11 | -1.577 | -1.66 | -0.346 | -0.245 | -0.816 |
| 08 | -3.578 | 0.031 | 0.443 | -1.0 | -0.626 | 1.147 | -0.617 | -1.626 | -0.624 | 0.701 | 1.549 | -2.123 | -1.646 | -0.471 | -0.983 | -3.578 |
| 09 | -0.805 | -0.239 | -3.578 | -3.578 | -0.346 | 1.801 | -3.578 | -3.578 | 0.802 | 1.554 | -1.664 | -3.578 | -1.687 | -0.952 | -2.178 | -3.578 |
| 10 | -3.578 | -3.578 | 1.226 | -2.24 | -0.817 | -1.59 | 2.372 | -0.51 | -1.66 | -3.578 | -2.185 | -3.578 | -3.578 | -3.578 | -3.578 | -3.578 |
| 11 | -3.578 | -3.578 | -0.504 | -3.578 | -3.578 | -3.578 | -1.59 | -3.578 | -3.578 | -3.578 | 2.646 | -2.209 | -3.578 | -1.651 | -0.664 | -3.578 |
| 12 | -3.578 | -3.578 | -3.578 | -3.578 | -1.651 | -3.578 | -3.578 | -3.578 | 2.675 | -3.578 | -0.164 | -3.578 | -3.578 | -3.578 | -2.209 | -3.578 |
| 13 | 2.621 | -2.214 | -0.472 | -1.263 | -3.578 | -3.578 | -3.578 | -3.578 | -0.376 | -3.578 | -1.302 | -3.578 | -3.578 | -3.578 | -3.578 | -3.578 |
| 14 | 0.539 | -1.642 | 2.53 | -3.578 | -3.578 | -3.578 | -2.214 | -3.578 | -1.238 | -2.179 | -0.652 | -3.578 | -3.578 | -3.578 | -1.263 | -3.578 |
| 15 | -1.028 | 0.143 | -0.823 | -2.207 | -3.578 | -1.642 | -3.578 | -2.179 | -0.153 | 1.496 | 0.604 | 1.843 | -3.578 | -3.578 | -3.578 | -3.578 |
| 16 | -0.627 | -2.203 | -0.815 | -1.689 | 0.188 | -0.033 | 1.055 | -0.234 | -0.773 | -1.622 | 0.322 | -1.253 | 0.187 | -0.32 | 1.505 | -2.049 |
| 17 | -0.002 | -1.172 | 0.599 | -1.272 | -2.165 | -0.469 | 0.055 | -1.585 | 0.341 | 0.51 | 1.702 | -0.442 | -2.252 | -0.966 | -0.132 | -3.578 |
| 18 | -0.125 | -0.092 | -0.339 | -2.137 | -1.585 | 0.15 | -0.12 | -0.318 | 1.238 | 0.807 | 0.988 | -0.15 | -3.578 | -1.299 | -0.19 | -3.578 |
| 19 | 0.812 | -0.473 | 0.345 | -1.279 | -0.789 | -0.013 | 0.856 | -0.23 | 0.552 | -0.473 | 0.905 | -1.269 | -0.575 | -2.132 | -0.249 | -1.292 |
| 20 | 0.389 | -0.623 | 1.029 | -1.641 | -0.609 | -0.335 | -0.028 | -2.123 | 0.156 | 0.997 | 0.891 | -0.359 | -2.202 | -1.643 | 0.116 | -1.604 |
| 21 | -0.331 | -0.23 | 0.342 | -0.977 | -1.035 | 0.465 | 0.398 | -0.331 | 0.044 | 0.855 | 0.929 | 0.34 | -2.141 | -0.609 | -0.647 | -3.578 |
| 22 | -1.052 | -0.958 | 0.2 | -1.268 | 0.408 | 0.063 | 0.201 | 0.398 | -0.041 | 0.438 | 0.93 | -0.094 | -0.907 | -0.136 | -0.329 | -0.632 |