Model info
| Transcription factor | EHF | ||||||||
| Model | EHF_HUMAN.H10DI.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 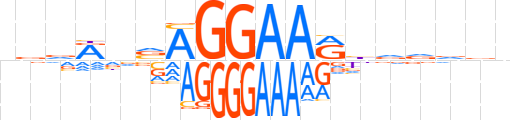 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 18 | ||||||||
| Quality | C | ||||||||
| Consensus | nvvRnvAGGAARbdddvn | ||||||||
| wAUC | 0.8322766701438603 | ||||||||
| Best AUC | 0.8322766701438604 | ||||||||
| Benchmark datasets | 1 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | Ets-related factors{3.5.2} | ||||||||
| TF subfamily | EHF-like factors{3.5.2.4} | ||||||||
| HGNC | 3246 | ||||||||
| EntrezGene | 26298 | ||||||||
| UniProt ID | EHF_HUMAN | ||||||||
| UniProt AC | Q9NZC4 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 32.754 | 23.409 | 67.097 | 6.977 | 33.829 | 29.232 | 26.2 | 31.992 | 29.702 | 21.57 | 87.061 | 13.753 | 24.356 | 18.513 | 33.148 | 20.406 |
| 02 | 35.974 | 13.088 | 50.267 | 21.313 | 37.463 | 21.608 | 13.634 | 20.018 | 92.627 | 30.422 | 83.378 | 7.079 | 20.199 | 18.803 | 28.354 | 5.772 |
| 03 | 136.257 | 4.125 | 28.15 | 17.732 | 58.093 | 4.707 | 4.348 | 16.774 | 128.603 | 12.974 | 23.732 | 10.324 | 26.296 | 8.594 | 4.984 | 14.308 |
| 04 | 109.803 | 91.696 | 94.054 | 53.695 | 17.68 | 9.818 | 0.935 | 1.966 | 29.903 | 8.652 | 19.465 | 3.195 | 11.929 | 15.879 | 15.035 | 16.295 |
| 05 | 44.742 | 62.962 | 59.613 | 1.998 | 33.625 | 81.645 | 7.904 | 2.87 | 19.999 | 37.796 | 69.606 | 2.088 | 26.986 | 9.095 | 38.099 | 0.972 |
| 06 | 109.748 | 6.204 | 9.4 | 0.0 | 149.761 | 39.629 | 2.109 | 0.0 | 148.097 | 21.453 | 5.672 | 0.0 | 5.059 | 0.0 | 2.87 | 0.0 |
| 07 | 0.0 | 0.0 | 411.678 | 0.986 | 0.0 | 0.0 | 67.286 | 0.0 | 0.0 | 0.0 | 20.05 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.9 | 3.072 | 490.042 | 0.0 | 0.0 | 0.0 | 0.986 | 0.0 |
| 09 | 5.9 | 0.0 | 0.0 | 0.0 | 3.072 | 0.0 | 0.0 | 0.0 | 483.568 | 0.906 | 3.717 | 2.837 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 482.733 | 1.054 | 3.812 | 4.942 | 0.906 | 0.0 | 0.0 | 0.0 | 3.717 | 0.0 | 0.0 | 0.0 | 1.875 | 0.0 | 0.0 | 0.962 |
| 11 | 176.375 | 23.275 | 287.691 | 1.889 | 0.0 | 0.0 | 1.054 | 0.0 | 1.933 | 0.0 | 1.879 | 0.0 | 2.978 | 0.0 | 2.926 | 0.0 |
| 12 | 38.462 | 38.238 | 29.985 | 74.601 | 2.049 | 7.826 | 0.912 | 12.488 | 39.389 | 57.592 | 51.022 | 145.547 | 1.004 | 0.0 | 0.0 | 0.886 |
| 13 | 36.214 | 9.898 | 25.966 | 8.826 | 37.59 | 15.993 | 31.009 | 19.064 | 30.268 | 17.829 | 27.665 | 6.157 | 68.595 | 16.082 | 113.768 | 35.076 |
| 14 | 54.524 | 16.958 | 84.762 | 16.423 | 31.004 | 7.084 | 12.981 | 8.733 | 65.743 | 19.879 | 84.792 | 27.993 | 18.182 | 10.064 | 29.672 | 11.205 |
| 15 | 71.074 | 13.636 | 64.002 | 20.74 | 22.62 | 6.159 | 10.003 | 15.203 | 81.337 | 30.581 | 78.013 | 22.277 | 12.262 | 11.771 | 17.371 | 22.95 |
| 16 | 62.749 | 19.793 | 84.367 | 20.385 | 16.663 | 22.641 | 8.221 | 14.621 | 57.066 | 31.083 | 62.42 | 18.822 | 16.634 | 11.087 | 29.045 | 24.404 |
| 17 | 58.028 | 12.897 | 67.746 | 14.441 | 29.844 | 22.852 | 8.353 | 23.554 | 67.239 | 41.03 | 55.528 | 20.256 | 11.623 | 20.133 | 23.37 | 23.106 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.046 | -0.285 | 0.758 | -1.458 | 0.078 | -0.066 | -0.174 | 0.023 | -0.05 | -0.365 | 1.017 | -0.805 | -0.246 | -0.515 | 0.058 | -0.42 |
| 02 | 0.139 | -0.853 | 0.471 | -0.377 | 0.179 | -0.363 | -0.814 | -0.439 | 1.078 | -0.027 | 0.974 | -1.444 | -0.43 | -0.5 | -0.096 | -1.636 |
| 03 | 1.463 | -1.947 | -0.103 | -0.557 | 0.614 | -1.826 | -1.899 | -0.612 | 1.405 | -0.862 | -0.271 | -1.083 | -0.17 | -1.259 | -1.773 | -0.767 |
| 04 | 1.248 | 1.068 | 1.094 | 0.536 | -0.56 | -1.131 | -3.174 | -2.598 | -0.044 | -1.253 | -0.466 | -2.178 | -0.943 | -0.665 | -0.719 | -0.64 |
| 05 | 0.355 | 0.694 | 0.64 | -2.584 | 0.072 | 0.953 | -1.339 | -2.273 | -0.439 | 0.188 | 0.794 | -2.547 | -0.145 | -1.205 | 0.196 | -3.147 |
| 06 | 1.247 | -1.568 | -1.173 | -4.4 | 1.557 | 0.235 | -2.539 | -4.4 | 1.546 | -0.371 | -1.653 | -4.4 | -1.759 | -4.4 | -2.273 | -4.4 |
| 07 | -4.4 | -4.4 | 2.567 | -3.136 | -4.4 | -4.4 | 0.76 | -4.4 | -4.4 | -4.4 | -0.437 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 |
| 08 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -1.616 | -2.213 | 2.741 | -4.4 | -4.4 | -4.4 | -3.136 | -4.4 |
| 09 | -1.616 | -4.4 | -4.4 | -4.4 | -2.213 | -4.4 | -4.4 | -4.4 | 2.728 | -3.197 | -2.042 | -2.283 | -4.4 | -4.4 | -4.4 | -4.4 |
| 10 | 2.726 | -3.088 | -2.019 | -1.781 | -3.197 | -4.4 | -4.4 | -4.4 | -2.042 | -4.4 | -4.4 | -4.4 | -2.637 | -4.4 | -4.4 | -3.154 |
| 11 | 1.72 | -0.29 | 2.209 | -2.631 | -4.4 | -4.4 | -3.088 | -4.4 | -2.612 | -4.4 | -2.636 | -4.4 | -2.241 | -4.4 | -2.256 | -4.4 |
| 12 | 0.205 | 0.2 | -0.041 | 0.863 | -2.563 | -1.348 | -3.192 | -0.899 | 0.229 | 0.606 | 0.485 | 1.529 | -3.124 | -4.4 | -4.4 | -3.212 |
| 13 | 0.146 | -1.124 | -0.183 | -1.234 | 0.183 | -0.658 | -0.008 | -0.486 | -0.032 | -0.552 | -0.12 | -1.576 | 0.779 | -0.653 | 1.283 | 0.114 |
| 14 | 0.551 | -0.601 | 0.99 | -0.632 | -0.008 | -1.443 | -0.861 | -1.244 | 0.737 | -0.445 | 0.99 | -0.109 | -0.533 | -1.107 | -0.051 | -1.004 |
| 15 | 0.815 | -0.814 | 0.711 | -0.404 | -0.319 | -1.575 | -1.113 | -0.708 | 0.949 | -0.021 | 0.907 | -0.334 | -0.917 | -0.956 | -0.577 | -0.304 |
| 16 | 0.691 | -0.45 | 0.985 | -0.421 | -0.618 | -0.318 | -1.301 | -0.746 | 0.597 | -0.005 | 0.686 | -0.499 | -0.62 | -1.014 | -0.072 | -0.244 |
| 17 | 0.613 | -0.868 | 0.767 | -0.758 | -0.045 | -0.308 | -1.286 | -0.279 | 0.76 | 0.269 | 0.569 | -0.427 | -0.969 | -0.433 | -0.286 | -0.298 |