Model info
| Transcription factor | Ctcf | ||||||||
| Model | CTCF_MOUSE.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 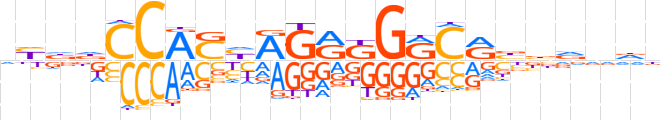 | ||||||||
| LOGO (reverse complement) | 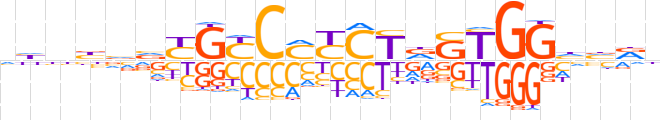 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Consensus | nYbdCCAShAGRKGRCRbbvndb | ||||||||
| wAUC | 0.9828675552675289 | ||||||||
| Best AUC | 0.9954072691558203 | ||||||||
| Benchmark datasets | 35 | ||||||||
| Aligned words | 502 | ||||||||
| TF family | More than 3 adjacent zinc finger factors{2.3.3} | ||||||||
| TF subfamily | CTCF-like factors{2.3.3.50} | ||||||||
| MGI | 109447 | ||||||||
| EntrezGene | 13018 | ||||||||
| UniProt ID | CTCF_MOUSE | ||||||||
| UniProt AC | Q61164 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 12.77 | 17.327 | 10.817 | 135.568 | 13.94 | 28.382 | 3.566 | 54.12 | 16.522 | 52.439 | 10.126 | 44.176 | 4.557 | 34.76 | 9.045 | 41.458 |
| 02 | 4.51 | 7.341 | 23.729 | 12.211 | 38.383 | 28.217 | 27.85 | 38.457 | 8.125 | 9.108 | 11.877 | 4.443 | 13.823 | 30.738 | 199.779 | 30.98 |
| 03 | 8.124 | 2.245 | 48.14 | 6.333 | 37.485 | 1.789 | 30.693 | 5.437 | 46.746 | 30.558 | 98.225 | 87.706 | 19.234 | 0.0 | 39.6 | 27.259 |
| 04 | 2.778 | 105.24 | 1.779 | 1.793 | 16.275 | 14.55 | 0.981 | 2.786 | 14.016 | 186.214 | 5.534 | 10.893 | 1.768 | 124.088 | 0.878 | 0.0 |
| 05 | 0.885 | 33.951 | 0.0 | 0.0 | 0.954 | 428.224 | 0.0 | 0.915 | 0.0 | 9.172 | 0.0 | 0.0 | 0.0 | 15.472 | 0.0 | 0.0 |
| 06 | 0.0 | 0.0 | 1.839 | 0.0 | 400.827 | 6.364 | 43.74 | 35.887 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.915 | 0.0 |
| 07 | 1.772 | 276.462 | 118.262 | 4.331 | 1.794 | 3.636 | 0.934 | 0.0 | 0.0 | 23.53 | 22.964 | 0.0 | 2.69 | 10.188 | 23.009 | 0.0 |
| 08 | 0.867 | 4.506 | 0.0 | 0.883 | 50.426 | 79.749 | 6.246 | 177.396 | 23.175 | 103.121 | 6.425 | 32.449 | 0.0 | 1.869 | 0.883 | 1.579 |
| 09 | 55.301 | 6.367 | 8.239 | 4.561 | 181.976 | 0.0 | 7.269 | 0.0 | 12.662 | 0.891 | 0.0 | 0.0 | 117.79 | 6.271 | 86.34 | 1.907 |
| 10 | 0.0 | 0.0 | 353.889 | 13.84 | 0.0 | 0.0 | 7.258 | 6.271 | 1.791 | 2.046 | 19.0 | 79.01 | 0.0 | 0.0 | 6.467 | 0.0 |
| 11 | 0.0 | 0.0 | 1.791 | 0.0 | 2.046 | 0.0 | 0.0 | 0.0 | 138.052 | 1.836 | 245.764 | 0.962 | 98.22 | 0.0 | 0.901 | 0.0 |
| 12 | 8.604 | 2.847 | 199.636 | 27.231 | 0.917 | 0.0 | 0.0 | 0.919 | 11.013 | 18.91 | 123.637 | 94.896 | 0.0 | 0.0 | 0.962 | 0.0 |
| 13 | 3.004 | 0.0 | 16.147 | 1.384 | 0.895 | 0.0 | 20.863 | 0.0 | 3.675 | 0.0 | 319.674 | 0.887 | 0.0 | 0.0 | 123.046 | 0.0 |
| 14 | 3.898 | 0.0 | 3.675 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 113.068 | 6.737 | 327.892 | 32.034 | 1.384 | 0.0 | 0.887 | 0.0 |
| 15 | 0.876 | 114.802 | 0.0 | 2.672 | 1.763 | 4.084 | 0.889 | 0.0 | 39.551 | 268.914 | 2.827 | 21.16 | 0.0 | 32.034 | 0.0 | 0.0 |
| 16 | 3.572 | 0.942 | 36.688 | 0.99 | 203.322 | 18.563 | 194.157 | 3.79 | 0.889 | 0.0 | 2.827 | 0.0 | 2.657 | 0.0 | 21.175 | 0.0 |
| 17 | 2.803 | 38.635 | 94.0 | 75.001 | 1.118 | 4.004 | 0.0 | 14.383 | 23.87 | 192.221 | 12.973 | 25.785 | 1.782 | 1.907 | 0.0 | 1.092 |
| 18 | 0.0 | 17.561 | 2.915 | 9.095 | 33.032 | 80.911 | 18.732 | 104.092 | 9.116 | 51.943 | 10.877 | 35.037 | 2.694 | 7.186 | 104.344 | 2.038 |
| 19 | 11.168 | 9.173 | 17.272 | 7.23 | 104.132 | 19.567 | 21.051 | 12.85 | 16.044 | 23.485 | 95.528 | 1.811 | 42.767 | 27.513 | 76.411 | 3.571 |
| 20 | 26.884 | 24.54 | 87.029 | 35.657 | 11.845 | 15.608 | 27.422 | 24.863 | 102.007 | 29.031 | 26.819 | 52.405 | 5.488 | 3.574 | 12.736 | 3.664 |
| 21 | 117.56 | 5.347 | 18.625 | 4.692 | 27.356 | 14.469 | 12.646 | 18.283 | 82.649 | 27.467 | 20.906 | 22.985 | 22.969 | 18.975 | 47.066 | 27.579 |
| 22 | 23.603 | 23.343 | 87.327 | 116.26 | 19.031 | 12.769 | 17.261 | 17.197 | 20.148 | 26.405 | 35.908 | 16.782 | 9.057 | 17.442 | 36.62 | 10.419 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.857 | -0.559 | -1.017 | 1.479 | -0.771 | -0.074 | -2.059 | 0.565 | -0.606 | 0.533 | -1.081 | 0.363 | -1.835 | 0.126 | -1.189 | 0.3 |
| 02 | -1.845 | -1.389 | -0.251 | -0.9 | 0.224 | -0.08 | -0.093 | 0.226 | -1.292 | -1.183 | -0.927 | -1.859 | -0.78 | 0.005 | 1.866 | 0.012 |
| 03 | -1.292 | -2.466 | 0.449 | -1.528 | 0.201 | -2.656 | 0.003 | -1.671 | 0.419 | -0.001 | 1.158 | 1.045 | -0.457 | -4.383 | 0.255 | -0.114 |
| 04 | -2.281 | 1.226 | -2.661 | -2.654 | -0.62 | -0.73 | -3.12 | -2.279 | -0.766 | 1.795 | -1.655 | -1.01 | -2.666 | 1.391 | -3.198 | -4.383 |
| 05 | -3.193 | 0.103 | -4.383 | -4.383 | -3.14 | 2.627 | -4.383 | -3.169 | -4.383 | -1.176 | -4.383 | -4.383 | -4.383 | -0.67 | -4.383 | -4.383 |
| 06 | -4.383 | -4.383 | -2.633 | -4.383 | 2.561 | -1.524 | 0.354 | 0.158 | -4.383 | -4.383 | -4.383 | -4.383 | -4.383 | -4.383 | -3.169 | -4.383 |
| 07 | -2.664 | 2.19 | 1.343 | -1.882 | -2.654 | -2.041 | -3.155 | -4.383 | -4.383 | -0.259 | -0.283 | -4.383 | -2.31 | -1.075 | -0.281 | -4.383 |
| 08 | -3.207 | -1.846 | -4.383 | -3.195 | 0.495 | 0.95 | -1.541 | 1.747 | -0.274 | 1.206 | -1.515 | 0.058 | -4.383 | -2.62 | -3.195 | -2.757 |
| 09 | 0.586 | -1.523 | -1.279 | -1.835 | 1.772 | -4.383 | -1.398 | -4.383 | -0.865 | -3.188 | -4.383 | -4.383 | 1.339 | -1.538 | 1.029 | -2.603 |
| 10 | -4.383 | -4.383 | 2.437 | -0.778 | -4.383 | -4.383 | -1.399 | -1.538 | -2.655 | -2.544 | -0.469 | 0.941 | -4.383 | -4.383 | -1.509 | -4.383 |
| 11 | -4.383 | -4.383 | -2.655 | -4.383 | -2.544 | -4.383 | -4.383 | -4.383 | 1.497 | -2.635 | 2.072 | -3.134 | 1.158 | -4.383 | -3.181 | -4.383 |
| 12 | -1.237 | -2.26 | 1.865 | -0.115 | -3.168 | -4.383 | -4.383 | -3.167 | -1.0 | -0.474 | 1.387 | 1.123 | -4.383 | -4.383 | -3.134 | -4.383 |
| 13 | -2.212 | -4.383 | -0.628 | -2.862 | -3.185 | -4.383 | -0.377 | -4.383 | -2.032 | -4.383 | 2.335 | -3.191 | -4.383 | -4.383 | 1.382 | -4.383 |
| 14 | -1.978 | -4.383 | -2.032 | -4.383 | -4.383 | -4.383 | -4.383 | -4.383 | 1.298 | -1.47 | 2.36 | 0.045 | -2.862 | -4.383 | -3.191 | -4.383 |
| 15 | -3.2 | 1.313 | -4.383 | -2.315 | -2.668 | -1.936 | -3.19 | -4.383 | 0.254 | 2.162 | -2.266 | -0.363 | -4.383 | 0.045 | -4.383 | -4.383 |
| 16 | -2.058 | -3.149 | 0.179 | -3.114 | 1.883 | -0.492 | 1.837 | -2.004 | -3.19 | -4.383 | -2.266 | -4.383 | -2.32 | -4.383 | -0.363 | -4.383 |
| 17 | -2.274 | 0.231 | 1.114 | 0.889 | -3.025 | -1.954 | -4.383 | -0.741 | -0.245 | 1.827 | -0.841 | -0.169 | -2.659 | -2.603 | -4.383 | -3.042 |
| 18 | -4.383 | -0.546 | -2.239 | -1.184 | 0.076 | 0.965 | -0.483 | 1.215 | -1.182 | 0.524 | -1.012 | 0.134 | -2.308 | -1.409 | 1.218 | -2.548 |
| 19 | -0.986 | -1.176 | -0.562 | -1.403 | 1.216 | -0.44 | -0.368 | -0.85 | -0.634 | -0.261 | 1.13 | -2.646 | 0.331 | -0.105 | 0.908 | -2.058 |
| 20 | -0.128 | -0.218 | 1.037 | 0.151 | -0.929 | -0.661 | -0.108 | -0.205 | 1.195 | -0.052 | -0.13 | 0.533 | -1.663 | -2.057 | -0.859 | -2.034 |
| 21 | 1.337 | -1.687 | -0.488 | -1.808 | -0.111 | -0.735 | -0.866 | -0.507 | 0.986 | -0.107 | -0.375 | -0.282 | -0.283 | -0.47 | 0.426 | -0.103 |
| 22 | -0.256 | -0.267 | 1.041 | 1.326 | -0.467 | -0.857 | -0.563 | -0.567 | -0.411 | -0.145 | 0.158 | -0.59 | -1.188 | -0.553 | 0.178 | -1.053 |