Model info
| Transcription factor | Rela | ||||||||
| Model | TF65_MOUSE.H10MO.C | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 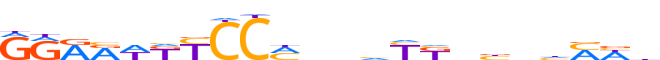 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 22 | ||||||||
| Quality | C | ||||||||
| Consensus | RRRRWKYCCMnnvYKdbnhMWb | ||||||||
| wAUC | 0.8139897406768539 | ||||||||
| Best AUC | 0.8139897406768538 | ||||||||
| Benchmark datasets | 1 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | NF-kappaB-related factors{6.1.1} | ||||||||
| TF subfamily | NF-kappaB p65 subunit-like factors{6.1.1.2} | ||||||||
| MGI | 103290 | ||||||||
| EntrezGene | 19697 | ||||||||
| UniProt ID | TF65_MOUSE | ||||||||
| UniProt AC | Q04207 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 64.45 | 11.996 | 391.171 | 32.383 |
| 02 | 80.513 | 3.783 | 368.039 | 47.665 |
| 03 | 324.247 | 21.922 | 140.764 | 13.067 |
| 04 | 327.486 | 72.945 | 77.86 | 21.709 |
| 05 | 203.085 | 25.288 | 23.897 | 247.731 |
| 06 | 49.436 | 42.869 | 75.61 | 332.085 |
| 07 | 11.019 | 123.13 | 26.95 | 338.901 |
| 08 | 20.137 | 450.528 | 4.972 | 24.363 |
| 09 | 12.887 | 458.35 | 0.0 | 28.763 |
| 10 | 197.072 | 233.879 | 13.628 | 55.422 |
| 11 | 141.539 | 109.24 | 121.091 | 128.13 |
| 12 | 146.563 | 70.515 | 132.546 | 150.376 |
| 13 | 203.771 | 125.736 | 115.877 | 54.615 |
| 14 | 58.837 | 65.401 | 23.665 | 352.096 |
| 15 | 33.713 | 39.282 | 122.903 | 304.102 |
| 16 | 102.883 | 66.777 | 173.525 | 156.815 |
| 17 | 62.244 | 260.681 | 88.262 | 88.813 |
| 18 | 114.826 | 123.371 | 135.779 | 126.024 |
| 19 | 139.453 | 191.981 | 36.646 | 131.921 |
| 20 | 277.583 | 179.191 | 28.419 | 14.806 |
| 21 | 311.554 | 59.67 | 58.383 | 70.392 |
| 22 | 60.914 | 137.994 | 108.088 | 193.004 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.651 | -2.234 | 1.132 | -1.316 |
| 02 | -0.433 | -3.166 | 1.072 | -0.944 |
| 03 | 0.946 | -1.685 | 0.117 | -2.158 |
| 04 | 0.956 | -0.53 | -0.466 | -1.694 |
| 05 | 0.481 | -1.551 | -1.604 | 0.678 |
| 06 | -0.909 | -1.047 | -0.495 | 0.969 |
| 07 | -2.309 | -0.015 | -1.491 | 0.99 |
| 08 | -1.764 | 1.273 | -2.965 | -1.586 |
| 09 | -2.171 | 1.29 | -4.4 | -1.429 |
| 10 | 0.451 | 0.621 | -2.121 | -0.798 |
| 11 | 0.123 | -0.133 | -0.031 | 0.024 |
| 12 | 0.157 | -0.563 | 0.058 | 0.183 |
| 13 | 0.484 | 0.006 | -0.075 | -0.812 |
| 14 | -0.74 | -0.637 | -1.613 | 1.028 |
| 15 | -1.278 | -1.131 | -0.017 | 0.882 |
| 16 | -0.192 | -0.616 | 0.325 | 0.224 |
| 17 | -0.685 | 0.729 | -0.343 | -0.337 |
| 18 | -0.084 | -0.013 | 0.082 | 0.008 |
| 19 | 0.108 | 0.425 | -1.198 | 0.053 |
| 20 | 0.791 | 0.356 | -1.44 | -2.046 |
| 21 | 0.906 | -0.726 | -0.747 | -0.565 |
| 22 | -0.706 | 0.098 | -0.143 | 0.43 |