Model info
| Transcription factor | Stat6 | ||||||||
| Model | STAT6_MOUSE.H10DI.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 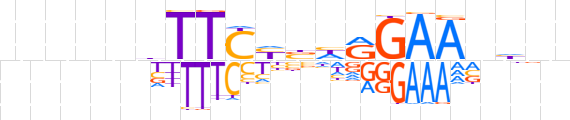 | ||||||||
| LOGO (reverse complement) | 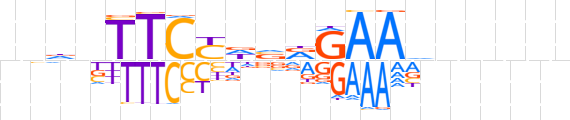 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 20 | ||||||||
| Quality | C | ||||||||
| Consensus | nnnnnbTTCYbhRGAAvhnn | ||||||||
| wAUC | 0.8262492362491791 | ||||||||
| Best AUC | 0.8262492362491792 | ||||||||
| Benchmark datasets | 1 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | STAT factors{6.2.1} | ||||||||
| TF subfamily | STAT6{6.2.1.0.7} | ||||||||
| MGI | 103034 | ||||||||
| EntrezGene | 20852 | ||||||||
| UniProt ID | STAT6_MOUSE | ||||||||
| UniProt AC | P52633 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 24.384 | 39.733 | 24.654 | 27.936 | 45.339 | 33.273 | 7.122 | 64.086 | 11.756 | 14.78 | 14.343 | 38.71 | 13.832 | 45.815 | 44.524 | 49.714 |
| 02 | 15.483 | 16.622 | 36.694 | 26.512 | 35.973 | 40.067 | 5.941 | 51.621 | 14.677 | 20.238 | 33.857 | 21.87 | 20.26 | 47.707 | 56.247 | 56.231 |
| 03 | 12.41 | 17.519 | 38.825 | 17.638 | 26.545 | 25.293 | 15.789 | 57.007 | 24.328 | 43.133 | 27.762 | 37.516 | 15.011 | 58.504 | 44.577 | 38.142 |
| 04 | 28.989 | 13.911 | 30.527 | 4.866 | 65.178 | 29.984 | 6.448 | 42.84 | 41.437 | 31.874 | 32.277 | 21.365 | 19.872 | 30.79 | 58.843 | 40.798 |
| 05 | 21.114 | 31.909 | 51.042 | 51.411 | 23.046 | 30.102 | 4.108 | 49.302 | 33.607 | 28.218 | 18.79 | 47.48 | 3.752 | 31.428 | 24.287 | 50.401 |
| 06 | 0.914 | 9.309 | 4.022 | 67.275 | 0.0 | 0.966 | 0.0 | 120.691 | 0.857 | 2.033 | 0.0 | 95.338 | 1.972 | 5.167 | 0.842 | 190.614 |
| 07 | 0.914 | 0.0 | 0.857 | 1.972 | 0.0 | 0.0 | 2.117 | 15.36 | 0.0 | 0.0 | 0.842 | 4.022 | 5.819 | 2.64 | 13.146 | 452.311 |
| 08 | 0.0 | 6.733 | 0.0 | 0.0 | 0.0 | 2.64 | 0.0 | 0.0 | 3.757 | 9.152 | 3.061 | 0.993 | 26.58 | 398.334 | 7.62 | 41.13 |
| 09 | 7.764 | 9.884 | 1.809 | 10.88 | 63.025 | 106.93 | 4.476 | 242.428 | 3.862 | 0.0 | 3.058 | 3.762 | 2.904 | 30.72 | 3.002 | 5.496 |
| 10 | 11.639 | 28.033 | 25.435 | 12.449 | 2.09 | 98.545 | 5.344 | 41.555 | 0.0 | 6.4 | 5.057 | 0.888 | 18.739 | 118.615 | 87.5 | 37.712 |
| 11 | 12.726 | 6.675 | 0.0 | 13.067 | 100.03 | 56.526 | 4.211 | 90.824 | 57.043 | 34.129 | 0.0 | 32.163 | 43.47 | 7.977 | 0.863 | 40.294 |
| 12 | 69.386 | 1.002 | 137.219 | 5.662 | 67.807 | 0.0 | 33.51 | 3.992 | 2.076 | 0.0 | 1.707 | 1.291 | 39.797 | 3.598 | 124.332 | 8.622 |
| 13 | 1.768 | 3.4 | 173.032 | 0.865 | 0.0 | 0.861 | 1.968 | 1.772 | 4.861 | 8.153 | 269.588 | 14.166 | 0.912 | 1.192 | 13.577 | 3.885 |
| 14 | 6.652 | 0.0 | 0.0 | 0.888 | 7.776 | 2.103 | 3.727 | 0.0 | 438.71 | 6.434 | 7.323 | 5.699 | 16.014 | 3.544 | 1.131 | 0.0 |
| 15 | 431.877 | 10.304 | 26.972 | 0.0 | 6.494 | 2.683 | 2.049 | 0.855 | 7.514 | 3.511 | 1.154 | 0.0 | 4.698 | 1.889 | 0.0 | 0.0 |
| 16 | 161.252 | 120.393 | 119.324 | 49.614 | 2.453 | 7.587 | 3.181 | 5.165 | 5.189 | 17.063 | 3.296 | 4.627 | 0.0 | 0.855 | 0.0 | 0.0 |
| 17 | 41.734 | 41.951 | 17.019 | 68.19 | 25.12 | 28.766 | 3.03 | 88.982 | 34.159 | 34.042 | 17.342 | 40.259 | 6.63 | 10.011 | 7.49 | 35.275 |
| 18 | 22.89 | 19.318 | 49.806 | 15.63 | 36.071 | 38.259 | 11.967 | 28.473 | 6.525 | 15.568 | 15.071 | 7.717 | 22.145 | 59.78 | 113.645 | 37.136 |
| 19 | 13.324 | 28.698 | 29.505 | 16.104 | 30.596 | 32.763 | 14.025 | 55.542 | 59.447 | 49.037 | 48.769 | 33.236 | 6.31 | 15.731 | 37.306 | 29.609 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.245 | 0.238 | -0.234 | -0.111 | 0.368 | 0.062 | -1.438 | 0.712 | -0.958 | -0.735 | -0.764 | 0.212 | -0.8 | 0.379 | 0.35 | 0.46 |
| 02 | -0.69 | -0.621 | 0.159 | -0.162 | 0.139 | 0.246 | -1.609 | 0.497 | -0.742 | -0.428 | 0.079 | -0.352 | -0.427 | 0.419 | 0.582 | 0.582 |
| 03 | -0.905 | -0.569 | 0.215 | -0.563 | -0.161 | -0.209 | -0.671 | 0.596 | -0.247 | 0.319 | -0.117 | 0.181 | -0.72 | 0.621 | 0.352 | 0.197 |
| 04 | -0.074 | -0.794 | -0.023 | -1.795 | 0.729 | -0.041 | -1.532 | 0.312 | 0.279 | 0.02 | 0.032 | -0.375 | -0.446 | -0.015 | 0.627 | 0.264 |
| 05 | -0.386 | 0.021 | 0.486 | 0.493 | -0.3 | -0.037 | -1.951 | 0.451 | 0.072 | -0.101 | -0.501 | 0.414 | -2.033 | 0.006 | -0.249 | 0.473 |
| 06 | -3.19 | -1.182 | -1.97 | 0.76 | -4.4 | -3.151 | -4.4 | 1.342 | -3.235 | -2.57 | -4.4 | 1.107 | -2.596 | -1.74 | -3.247 | 1.798 |
| 07 | -3.19 | -4.4 | -3.235 | -2.596 | -4.4 | -4.4 | -2.536 | -0.698 | -4.4 | -4.4 | -3.247 | -1.97 | -1.629 | -2.346 | -0.849 | 2.661 |
| 08 | -4.4 | -1.491 | -4.4 | -4.4 | -4.4 | -2.346 | -4.4 | -4.4 | -2.032 | -1.199 | -2.216 | -3.132 | -0.16 | 2.534 | -1.374 | 0.272 |
| 09 | -1.356 | -1.125 | -2.667 | -1.032 | 0.695 | 1.221 | -1.872 | 2.038 | -2.007 | -4.4 | -2.217 | -2.031 | -2.263 | -0.017 | -2.233 | -1.682 |
| 10 | -0.967 | -0.107 | -0.203 | -0.902 | -2.547 | 1.14 | -1.708 | 0.282 | -4.4 | -1.539 | -1.76 | -3.21 | -0.503 | 1.325 | 1.022 | 0.186 |
| 11 | -0.881 | -1.499 | -4.4 | -0.855 | 1.155 | 0.587 | -1.928 | 1.059 | 0.596 | 0.087 | -4.4 | 0.028 | 0.327 | -1.33 | -3.23 | 0.251 |
| 12 | 0.791 | -3.125 | 1.47 | -1.654 | 0.768 | -4.4 | 0.069 | -1.977 | -2.552 | -4.4 | -2.714 | -2.936 | 0.239 | -2.071 | 1.372 | -1.256 |
| 13 | -2.686 | -2.122 | 1.701 | -3.229 | -4.4 | -3.232 | -2.597 | -2.684 | -1.796 | -1.309 | 2.144 | -0.776 | -3.192 | -2.997 | -0.818 | -2.002 |
| 14 | -1.503 | -4.4 | -4.4 | -3.21 | -1.355 | -2.541 | -2.04 | -4.4 | 2.63 | -1.534 | -1.412 | -1.648 | -0.657 | -2.085 | -3.036 | -4.4 |
| 15 | 2.615 | -1.085 | -0.145 | -4.4 | -1.525 | -2.332 | -2.564 | -3.237 | -1.387 | -2.093 | -3.021 | -4.4 | -1.828 | -2.631 | -4.4 | -4.4 |
| 16 | 1.631 | 1.34 | 1.331 | 0.458 | -2.41 | -1.378 | -2.182 | -1.74 | -1.736 | -0.595 | -2.15 | -1.842 | -4.4 | -3.237 | -4.4 | -4.4 |
| 17 | 0.286 | 0.291 | -0.597 | 0.774 | -0.215 | -0.082 | -2.225 | 1.038 | 0.088 | 0.085 | -0.579 | 0.251 | -1.506 | -1.113 | -1.39 | 0.12 |
| 18 | -0.307 | -0.473 | 0.462 | -0.681 | 0.142 | 0.2 | -0.94 | -0.092 | -1.521 | -0.685 | -0.716 | -1.362 | -0.339 | 0.643 | 1.282 | 0.171 |
| 19 | -0.836 | -0.084 | -0.057 | -0.652 | -0.021 | 0.047 | -0.786 | 0.57 | 0.637 | 0.446 | 0.441 | 0.061 | -1.553 | -0.674 | 0.175 | -0.053 |