Model info
| Transcription factor | STAT2 | ||||||||
| Model | STAT2_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 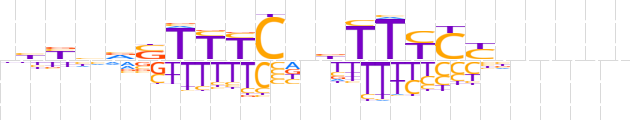 | ||||||||
| LOGO (reverse complement) | 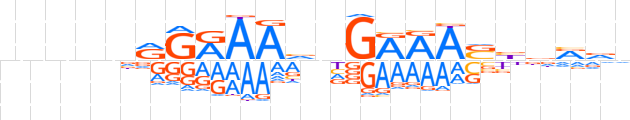 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 22 | ||||||||
| Quality | A | ||||||||
| Consensus | nbYhvSTTYCnbTTYCYnnnnn | ||||||||
| wAUC | 0.8691216148560169 | ||||||||
| Best AUC | 0.9238808701755828 | ||||||||
| Benchmark datasets | 2 | ||||||||
| Aligned words | 416 | ||||||||
| TF family | STAT factors{6.2.1} | ||||||||
| TF subfamily | STAT2{6.2.1.0.2} | ||||||||
| HGNC | 11363 | ||||||||
| EntrezGene | 6773 | ||||||||
| UniProt ID | STAT2_HUMAN | ||||||||
| UniProt AC | P52630 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 19.237 | 7.39 | 11.259 | 34.272 | 14.113 | 42.638 | 9.678 | 58.437 | 12.453 | 14.807 | 10.557 | 32.598 | 5.533 | 37.818 | 23.302 | 78.908 |
| 02 | 2.091 | 2.378 | 18.959 | 27.909 | 10.513 | 16.922 | 4.388 | 70.829 | 5.945 | 14.295 | 4.966 | 29.59 | 6.742 | 35.356 | 39.84 | 122.279 |
| 03 | 7.068 | 10.332 | 3.306 | 4.584 | 11.167 | 23.625 | 3.252 | 30.906 | 14.689 | 34.456 | 6.285 | 12.723 | 60.16 | 87.275 | 35.004 | 68.169 |
| 04 | 55.981 | 3.072 | 23.857 | 10.174 | 98.11 | 33.475 | 9.51 | 14.592 | 17.906 | 6.054 | 17.522 | 6.364 | 38.324 | 11.173 | 45.599 | 21.288 |
| 05 | 4.818 | 73.845 | 124.767 | 6.892 | 2.794 | 44.998 | 3.673 | 2.308 | 3.447 | 24.679 | 61.709 | 6.653 | 6.183 | 21.627 | 13.378 | 11.231 |
| 06 | 1.026 | 0.0 | 2.9 | 13.315 | 12.729 | 11.6 | 3.169 | 137.65 | 10.213 | 6.622 | 4.823 | 181.87 | 0.0 | 3.85 | 0.844 | 22.389 |
| 07 | 0.0 | 9.426 | 6.356 | 8.186 | 0.894 | 5.398 | 0.0 | 15.78 | 0.791 | 6.71 | 0.736 | 3.499 | 3.69 | 49.488 | 11.348 | 290.699 |
| 08 | 0.0 | 0.0 | 0.0 | 5.375 | 4.675 | 35.625 | 2.08 | 28.641 | 2.729 | 3.473 | 1.571 | 10.667 | 5.699 | 30.666 | 13.176 | 268.622 |
| 09 | 1.82 | 10.213 | 0.0 | 1.071 | 1.187 | 59.589 | 0.0 | 8.989 | 0.0 | 16.828 | 0.0 | 0.0 | 1.541 | 289.838 | 0.781 | 21.145 |
| 10 | 0.781 | 0.0 | 1.706 | 2.06 | 118.438 | 81.961 | 90.243 | 85.826 | 0.781 | 0.0 | 0.0 | 0.0 | 7.064 | 8.485 | 4.863 | 10.793 |
| 11 | 4.65 | 28.907 | 29.431 | 64.074 | 17.226 | 19.49 | 1.839 | 51.891 | 18.137 | 18.35 | 16.224 | 44.101 | 9.506 | 25.013 | 20.464 | 43.696 |
| 12 | 0.0 | 4.742 | 0.791 | 43.985 | 1.17 | 14.462 | 0.0 | 76.129 | 2.127 | 10.657 | 0.0 | 55.174 | 1.114 | 30.82 | 0.0 | 171.828 |
| 13 | 1.17 | 0.0 | 0.0 | 3.241 | 3.846 | 3.468 | 0.76 | 52.607 | 0.0 | 0.0 | 0.0 | 0.791 | 12.744 | 13.859 | 0.985 | 319.528 |
| 14 | 0.0 | 5.269 | 0.0 | 12.491 | 0.0 | 5.267 | 0.0 | 12.06 | 0.985 | 0.0 | 0.0 | 0.76 | 2.607 | 162.578 | 2.105 | 208.879 |
| 15 | 0.0 | 3.591 | 0.0 | 0.0 | 0.836 | 123.066 | 1.488 | 47.724 | 0.0 | 0.0 | 0.0 | 2.105 | 4.98 | 194.204 | 3.498 | 31.507 |
| 16 | 1.139 | 2.968 | 0.836 | 0.874 | 15.093 | 194.276 | 6.728 | 104.764 | 1.685 | 2.382 | 0.0 | 0.919 | 2.098 | 36.564 | 14.887 | 27.787 |
| 17 | 1.885 | 4.047 | 9.932 | 4.151 | 40.582 | 87.387 | 26.636 | 81.585 | 0.852 | 5.995 | 10.929 | 4.675 | 11.007 | 46.263 | 47.179 | 29.894 |
| 18 | 12.4 | 6.882 | 27.26 | 7.783 | 43.548 | 44.922 | 20.955 | 34.267 | 14.076 | 20.557 | 42.767 | 17.276 | 14.773 | 20.174 | 40.773 | 44.586 |
| 19 | 25.623 | 10.59 | 28.566 | 20.018 | 22.438 | 25.751 | 4.024 | 40.322 | 40.978 | 25.684 | 43.088 | 22.006 | 11.902 | 18.443 | 14.328 | 59.24 |
| 20 | 55.514 | 8.53 | 15.741 | 21.154 | 18.84 | 24.581 | 3.15 | 33.896 | 31.24 | 33.63 | 13.51 | 11.626 | 19.117 | 44.997 | 25.17 | 52.302 |
| 21 | 65.022 | 5.962 | 41.378 | 12.35 | 16.152 | 42.82 | 12.903 | 39.864 | 13.649 | 16.938 | 12.852 | 14.133 | 20.598 | 38.293 | 23.778 | 36.309 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.289 | -1.216 | -0.811 | 0.28 | -0.592 | 0.496 | -0.957 | 0.809 | -0.714 | -0.545 | -0.873 | 0.23 | -1.489 | 0.377 | -0.101 | 1.108 |
| 02 | -2.362 | -2.252 | -0.303 | 0.077 | -0.878 | -0.415 | -1.704 | 1.0 | -1.421 | -0.579 | -1.59 | 0.135 | -1.303 | 0.311 | 0.429 | 1.544 |
| 03 | -1.258 | -0.894 | -1.962 | -1.664 | -0.819 | -0.087 | -1.977 | 0.178 | -0.553 | 0.285 | -1.369 | -0.693 | 0.838 | 1.208 | 0.301 | 0.962 |
| 04 | 0.766 | -2.028 | -0.078 | -0.909 | 1.325 | 0.257 | -0.974 | -0.559 | -0.359 | -1.404 | -0.381 | -1.357 | 0.391 | -0.819 | 0.563 | -0.19 |
| 05 | -1.618 | 1.042 | 1.564 | -1.282 | -2.111 | 0.55 | -1.867 | -2.278 | -1.924 | -0.044 | 0.863 | -1.315 | -1.384 | -0.174 | -0.644 | -0.814 |
| 06 | -2.927 | -4.242 | -2.079 | -0.649 | -0.692 | -0.782 | -2.0 | 1.662 | -0.906 | -1.32 | -1.617 | 1.94 | -4.242 | -1.824 | -3.066 | -0.14 |
| 07 | -4.242 | -0.983 | -1.358 | -1.118 | -3.026 | -1.512 | -4.242 | -0.483 | -3.11 | -1.307 | -3.159 | -1.911 | -1.863 | 0.644 | -0.804 | 2.408 |
| 08 | -4.242 | -4.242 | -4.242 | -1.516 | -1.646 | 0.318 | -2.367 | 0.103 | -2.132 | -1.918 | -2.599 | -0.864 | -1.461 | 0.17 | -0.659 | 2.329 |
| 09 | -2.479 | -0.905 | -4.242 | -2.896 | -2.819 | 0.828 | -4.242 | -1.028 | -4.242 | -0.42 | -4.242 | -4.242 | -2.615 | 2.405 | -3.119 | -0.196 |
| 10 | -3.119 | -4.242 | -2.532 | -2.375 | 1.512 | 1.145 | 1.241 | 1.191 | -3.119 | -4.242 | -4.242 | -4.242 | -1.258 | -1.084 | -1.609 | -0.852 |
| 11 | -1.651 | 0.112 | 0.129 | 0.901 | -0.397 | -0.276 | -2.47 | 0.691 | -0.347 | -0.335 | -0.456 | 0.53 | -0.975 | -0.031 | -0.228 | 0.521 |
| 12 | -4.242 | -1.632 | -3.11 | 0.527 | -2.829 | -0.568 | -4.242 | 1.072 | -2.347 | -0.864 | -4.242 | 0.752 | -2.866 | 0.175 | -4.242 | 1.883 |
| 13 | -2.829 | -4.242 | -4.242 | -1.979 | -1.825 | -1.919 | -3.138 | 0.705 | -4.242 | -4.242 | -4.242 | -3.11 | -0.691 | -0.61 | -2.957 | 2.503 |
| 14 | -4.242 | -1.534 | -4.242 | -0.711 | -4.242 | -1.535 | -4.242 | -0.745 | -2.957 | -4.242 | -4.242 | -3.138 | -2.172 | 1.828 | -2.357 | 2.078 |
| 15 | -4.242 | -1.887 | -4.242 | -4.242 | -3.073 | 1.55 | -2.642 | 0.608 | -4.242 | -4.242 | -4.242 | -2.357 | -1.587 | 2.006 | -1.911 | 0.197 |
| 16 | -2.85 | -2.058 | -3.073 | -3.042 | -0.526 | 2.006 | -1.305 | 1.39 | -2.542 | -2.251 | -4.242 | -3.007 | -2.359 | 0.344 | -0.54 | 0.073 |
| 17 | -2.449 | -1.778 | -0.932 | -1.755 | 0.447 | 1.209 | 0.031 | 1.141 | -3.06 | -1.413 | -0.84 | -1.646 | -0.833 | 0.577 | 0.597 | 0.145 |
| 18 | -0.718 | -1.283 | 0.054 | -1.166 | 0.517 | 0.548 | -0.205 | 0.28 | -0.594 | -0.224 | 0.499 | -0.394 | -0.547 | -0.242 | 0.452 | 0.54 |
| 19 | -0.007 | -0.871 | 0.1 | -0.25 | -0.138 | -0.002 | -1.784 | 0.441 | 0.457 | -0.005 | 0.507 | -0.157 | -0.758 | -0.33 | -0.577 | 0.823 |
| 20 | 0.758 | -1.079 | -0.485 | -0.196 | -0.31 | -0.048 | -2.005 | 0.269 | 0.188 | 0.261 | -0.634 | -0.78 | -0.295 | 0.55 | -0.025 | 0.699 |
| 21 | 0.915 | -1.419 | 0.466 | -0.722 | -0.46 | 0.5 | -0.679 | 0.43 | -0.624 | -0.414 | -0.683 | -0.591 | -0.222 | 0.39 | -0.081 | 0.337 |