Model info
| Transcription factor | Spi1 | ||||||||
| Model | SPI1_MOUSE.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 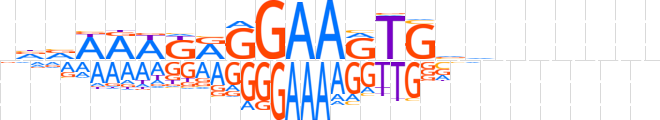 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Consensus | nvvWRWKRGGAARTGvvnnnnnn | ||||||||
| wAUC | 0.9587865003775525 | ||||||||
| Best AUC | 0.9891271635038471 | ||||||||
| Benchmark datasets | 7 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | Ets-related factors{3.5.2} | ||||||||
| TF subfamily | Spi-like factors{3.5.2.5} | ||||||||
| MGI | 98282 | ||||||||
| EntrezGene | 20375 | ||||||||
| UniProt ID | SPI1_MOUSE | ||||||||
| UniProt AC | P17433 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 48.748 | 13.399 | 40.597 | 5.812 | 75.31 | 12.341 | 9.629 | 21.439 | 108.138 | 15.358 | 17.379 | 17.208 | 34.564 | 23.942 | 26.299 | 19.165 |
| 02 | 142.257 | 19.311 | 92.622 | 12.57 | 38.338 | 12.397 | 5.793 | 8.513 | 51.919 | 6.631 | 25.677 | 9.676 | 26.349 | 8.666 | 21.934 | 6.676 |
| 03 | 220.232 | 6.712 | 21.417 | 10.501 | 32.774 | 1.897 | 0.94 | 11.394 | 117.157 | 4.161 | 11.489 | 13.219 | 14.392 | 5.701 | 2.86 | 14.481 |
| 04 | 327.751 | 3.874 | 42.346 | 10.585 | 14.672 | 2.786 | 0.0 | 1.013 | 23.048 | 3.807 | 9.851 | 0.0 | 17.153 | 0.0 | 13.433 | 19.009 |
| 05 | 298.816 | 0.974 | 20.291 | 62.543 | 8.598 | 0.925 | 0.0 | 0.944 | 42.613 | 0.941 | 16.301 | 5.774 | 4.847 | 0.0 | 0.988 | 24.772 |
| 06 | 21.934 | 19.969 | 243.673 | 69.297 | 0.0 | 0.941 | 0.925 | 0.974 | 2.914 | 0.0 | 34.667 | 0.0 | 8.724 | 0.0 | 85.309 | 0.0 |
| 07 | 18.252 | 0.93 | 14.39 | 0.0 | 19.916 | 0.0 | 0.994 | 0.0 | 274.699 | 24.93 | 64.006 | 0.94 | 0.973 | 0.0 | 66.566 | 2.732 |
| 08 | 0.997 | 0.0 | 312.844 | 0.0 | 16.459 | 0.0 | 9.401 | 0.0 | 74.366 | 0.0 | 71.59 | 0.0 | 2.732 | 0.0 | 0.94 | 0.0 |
| 09 | 0.0 | 0.0 | 93.196 | 1.357 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 394.775 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 483.784 | 1.953 | 2.234 | 0.0 | 1.357 | 0.0 | 0.0 | 0.0 |
| 11 | 484.165 | 0.0 | 0.0 | 0.976 | 1.953 | 0.0 | 0.0 | 0.0 | 2.234 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 12 | 81.114 | 46.577 | 360.661 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.976 | 0.0 |
| 13 | 0.0 | 0.972 | 0.0 | 80.142 | 2.931 | 0.0 | 0.0 | 43.646 | 8.534 | 10.399 | 10.566 | 332.138 | 0.0 | 0.0 | 0.0 | 0.0 |
| 14 | 0.0 | 0.951 | 10.514 | 0.0 | 1.877 | 5.662 | 2.891 | 0.942 | 0.0 | 3.759 | 6.807 | 0.0 | 10.483 | 38.939 | 405.559 | 0.944 |
| 15 | 1.013 | 4.685 | 6.662 | 0.0 | 18.996 | 10.408 | 3.849 | 16.057 | 112.534 | 155.501 | 127.889 | 29.847 | 0.0 | 0.942 | 0.944 | 0.0 |
| 16 | 57.562 | 16.092 | 46.48 | 12.409 | 96.595 | 30.535 | 5.711 | 38.696 | 37.359 | 31.933 | 39.371 | 30.681 | 4.723 | 8.624 | 28.624 | 3.933 |
| 17 | 69.166 | 79.812 | 20.102 | 27.16 | 22.923 | 20.982 | 3.933 | 39.347 | 26.81 | 33.012 | 36.565 | 23.799 | 15.302 | 21.214 | 31.002 | 18.201 |
| 18 | 38.313 | 32.766 | 45.869 | 17.253 | 99.195 | 16.121 | 3.867 | 35.835 | 31.125 | 18.995 | 23.113 | 18.369 | 15.31 | 33.843 | 33.299 | 26.053 |
| 19 | 27.682 | 82.776 | 55.206 | 18.28 | 28.142 | 26.99 | 5.65 | 40.943 | 21.992 | 23.052 | 30.541 | 30.564 | 18.214 | 19.25 | 33.898 | 26.148 |
| 20 | 24.716 | 18.36 | 34.657 | 18.297 | 31.955 | 19.015 | 3.858 | 97.24 | 39.566 | 25.018 | 35.602 | 25.109 | 16.36 | 27.793 | 45.158 | 26.624 |
| 21 | 29.794 | 16.396 | 54.058 | 12.349 | 26.028 | 20.104 | 5.74 | 38.315 | 33.674 | 27.038 | 29.865 | 28.697 | 21.29 | 18.125 | 96.2 | 31.655 |
| 22 | 33.997 | 24.835 | 30.862 | 21.092 | 21.189 | 16.267 | 6.703 | 37.504 | 30.241 | 29.874 | 44.367 | 81.382 | 12.355 | 24.245 | 38.192 | 36.223 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.462 | -0.809 | 0.28 | -1.609 | 0.894 | -0.889 | -1.129 | -0.35 | 1.254 | -0.676 | -0.556 | -0.565 | 0.121 | -0.241 | -0.149 | -0.46 |
| 02 | 1.527 | -0.452 | 1.1 | -0.871 | 0.223 | -0.885 | -1.612 | -1.247 | 0.524 | -1.485 | -0.172 | -1.124 | -0.147 | -1.23 | -0.328 | -1.478 |
| 03 | 1.963 | -1.473 | -0.351 | -1.045 | 0.068 | -2.607 | -3.15 | -0.967 | 1.334 | -1.918 | -0.958 | -0.822 | -0.74 | -1.627 | -2.255 | -0.734 |
| 04 | 2.36 | -1.984 | 0.322 | -1.038 | -0.721 | -2.278 | -4.382 | -3.096 | -0.279 | -1.999 | -1.107 | -4.382 | -0.569 | -4.382 | -0.807 | -0.468 |
| 05 | 2.268 | -3.125 | -0.404 | 0.709 | -1.237 | -3.161 | -4.382 | -3.147 | 0.328 | -3.149 | -0.618 | -1.615 | -1.778 | -4.382 | -3.114 | -0.208 |
| 06 | -0.327 | -0.42 | 2.064 | 0.811 | -4.382 | -3.149 | -3.161 | -3.125 | -2.239 | -4.382 | 0.124 | -4.382 | -1.224 | -4.382 | 1.018 | -4.382 |
| 07 | -0.508 | -3.158 | -0.74 | -4.382 | -0.422 | -4.382 | -3.11 | -4.382 | 2.184 | -0.202 | 0.732 | -3.15 | -3.125 | -4.382 | 0.771 | -2.296 |
| 08 | -3.108 | -4.382 | 2.314 | -4.382 | -0.609 | -4.382 | -1.152 | -4.382 | 0.881 | -4.382 | 0.843 | -4.382 | -2.296 | -4.382 | -3.15 | -4.382 |
| 09 | -4.382 | -4.382 | 1.106 | -2.877 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 | 2.546 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 |
| 10 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 | 2.749 | -2.583 | -2.469 | -4.382 | -2.877 | -4.382 | -4.382 | -4.382 |
| 11 | 2.75 | -4.382 | -4.382 | -3.123 | -2.583 | -4.382 | -4.382 | -4.382 | -2.469 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 |
| 12 | 0.968 | 0.416 | 2.456 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 | -4.382 | -3.123 | -4.382 |
| 13 | -4.382 | -3.126 | -4.382 | 0.956 | -2.234 | -4.382 | -4.382 | 0.352 | -1.245 | -1.055 | -1.039 | 2.374 | -4.382 | -4.382 | -4.382 | -4.382 |
| 14 | -4.382 | -3.142 | -1.044 | -4.382 | -2.616 | -1.633 | -2.246 | -3.149 | -4.382 | -2.011 | -1.46 | -4.382 | -1.047 | 0.239 | 2.573 | -3.147 |
| 15 | -3.096 | -1.809 | -1.48 | -4.382 | -0.469 | -1.054 | -1.989 | -0.633 | 1.294 | 1.616 | 1.421 | -0.024 | -4.382 | -3.149 | -3.147 | -4.382 |
| 16 | 0.627 | -0.631 | 0.414 | -0.884 | 1.142 | -0.002 | -1.625 | 0.233 | 0.198 | 0.043 | 0.25 | 0.003 | -1.802 | -1.235 | -0.065 | -1.97 |
| 17 | 0.809 | 0.951 | -0.413 | -0.117 | -0.284 | -0.371 | -1.97 | 0.249 | -0.13 | 0.075 | 0.177 | -0.247 | -0.68 | -0.36 | 0.013 | -0.511 |
| 18 | 0.223 | 0.068 | 0.401 | -0.563 | 1.168 | -0.629 | -1.985 | 0.157 | 0.017 | -0.469 | -0.276 | -0.502 | -0.68 | 0.1 | 0.084 | -0.158 |
| 19 | -0.098 | 0.988 | 0.585 | -0.506 | -0.082 | -0.123 | -1.635 | 0.289 | -0.325 | -0.279 | -0.001 | -0.001 | -0.51 | -0.456 | 0.102 | -0.155 |
| 20 | -0.21 | -0.502 | 0.124 | -0.505 | 0.043 | -0.468 | -1.987 | 1.148 | 0.255 | -0.198 | 0.15 | -0.194 | -0.615 | -0.094 | 0.386 | -0.137 |
| 21 | -0.026 | -0.613 | 0.564 | -0.889 | -0.159 | -0.413 | -1.62 | 0.223 | 0.095 | -0.122 | -0.023 | -0.063 | -0.357 | -0.515 | 1.137 | 0.034 |
| 22 | 0.105 | -0.205 | 0.009 | -0.366 | -0.361 | -0.62 | -1.474 | 0.202 | -0.011 | -0.023 | 0.368 | 0.971 | -0.888 | -0.229 | 0.22 | 0.167 |