Model info
| Transcription factor | SPI1 | ||||||||
| Model | SPI1_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 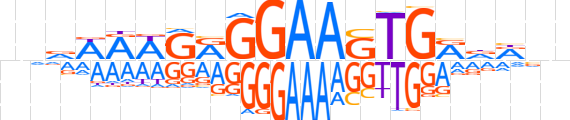 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 20 | ||||||||
| Quality | A | ||||||||
| Consensus | nvvWRWGRGGAAGTGRdRnn | ||||||||
| wAUC | 0.9871692881997829 | ||||||||
| Best AUC | 0.9944986702154056 | ||||||||
| Benchmark datasets | 9 | ||||||||
| Aligned words | 480 | ||||||||
| TF family | Ets-related factors{3.5.2} | ||||||||
| TF subfamily | Spi-like factors{3.5.2.5} | ||||||||
| HGNC | 11241 | ||||||||
| EntrezGene | 6688 | ||||||||
| UniProt ID | SPI1_HUMAN | ||||||||
| UniProt AC | P17947 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 52.472 | 29.312 | 53.416 | 20.395 | 57.905 | 19.573 | 2.204 | 24.987 | 45.817 | 24.227 | 29.215 | 16.606 | 31.958 | 12.88 | 37.209 | 22.195 |
| 02 | 95.69 | 11.383 | 69.451 | 11.628 | 55.889 | 10.741 | 9.958 | 9.404 | 61.934 | 11.358 | 41.811 | 6.941 | 23.446 | 11.199 | 36.746 | 12.792 |
| 03 | 194.244 | 0.0 | 21.023 | 21.691 | 28.477 | 5.891 | 1.116 | 9.196 | 114.921 | 9.029 | 20.39 | 13.626 | 14.821 | 3.069 | 7.845 | 15.029 |
| 04 | 308.398 | 3.875 | 27.288 | 12.902 | 13.037 | 0.871 | 2.035 | 2.046 | 36.677 | 1.779 | 11.041 | 0.877 | 12.346 | 1.122 | 12.505 | 33.57 |
| 05 | 289.952 | 4.504 | 20.371 | 55.631 | 7.647 | 0.0 | 0.0 | 0.0 | 31.187 | 1.014 | 9.477 | 11.191 | 8.616 | 0.854 | 0.865 | 39.061 |
| 06 | 44.626 | 14.659 | 273.178 | 4.939 | 4.596 | 0.919 | 0.857 | 0.0 | 4.199 | 0.0 | 24.575 | 1.939 | 12.546 | 2.741 | 89.744 | 0.853 |
| 07 | 22.561 | 1.81 | 41.595 | 0.0 | 16.503 | 0.0 | 1.816 | 0.0 | 258.238 | 33.981 | 94.194 | 1.94 | 1.936 | 0.0 | 5.794 | 0.0 |
| 08 | 8.239 | 0.886 | 290.114 | 0.0 | 30.768 | 0.0 | 5.023 | 0.0 | 8.866 | 0.0 | 134.533 | 0.0 | 0.0 | 0.0 | 1.94 | 0.0 |
| 09 | 0.0 | 0.0 | 47.873 | 0.0 | 0.0 | 0.0 | 0.886 | 0.0 | 0.0 | 0.0 | 431.61 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 480.369 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 11 | 476.267 | 0.0 | 0.0 | 4.102 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 12 | 8.465 | 140.022 | 327.78 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.001 | 0.0 | 3.102 | 0.0 |
| 13 | 0.0 | 0.89 | 0.0 | 8.576 | 7.618 | 6.548 | 0.0 | 125.856 | 2.921 | 2.772 | 6.106 | 319.082 | 0.0 | 0.0 | 0.0 | 0.0 |
| 14 | 0.0 | 0.0 | 10.54 | 0.0 | 0.865 | 0.0 | 9.344 | 0.0 | 0.0 | 0.0 | 6.106 | 0.0 | 1.043 | 14.984 | 437.486 | 0.0 |
| 15 | 1.043 | 0.0 | 0.865 | 0.0 | 7.605 | 2.155 | 2.287 | 2.938 | 310.881 | 35.081 | 106.789 | 10.725 | 0.0 | 0.0 | 0.0 | 0.0 |
| 16 | 194.912 | 10.767 | 98.528 | 15.323 | 11.677 | 2.999 | 4.014 | 18.545 | 24.734 | 22.828 | 48.939 | 13.439 | 3.761 | 0.902 | 8.135 | 0.866 |
| 17 | 192.025 | 9.401 | 26.826 | 6.831 | 13.079 | 11.595 | 1.056 | 11.766 | 97.977 | 17.926 | 21.555 | 22.158 | 7.486 | 14.637 | 12.59 | 13.461 |
| 18 | 67.635 | 111.32 | 98.285 | 33.327 | 19.105 | 19.194 | 3.073 | 12.189 | 26.755 | 17.128 | 12.959 | 5.186 | 12.702 | 13.672 | 9.533 | 18.309 |
| 19 | 48.128 | 24.811 | 30.782 | 22.475 | 45.347 | 37.566 | 7.158 | 71.242 | 39.032 | 20.938 | 25.815 | 38.065 | 7.752 | 17.204 | 13.775 | 30.278 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.553 | -0.024 | 0.571 | -0.381 | 0.651 | -0.421 | -2.463 | -0.181 | 0.418 | -0.211 | -0.027 | -0.582 | 0.062 | -0.83 | 0.212 | -0.298 |
| 02 | 1.15 | -0.949 | 0.831 | -0.929 | 0.616 | -1.005 | -1.078 | -1.133 | 0.718 | -0.951 | 0.328 | -1.423 | -0.244 | -0.965 | 0.2 | -0.836 |
| 03 | 1.856 | -4.367 | -0.351 | -0.32 | -0.052 | -1.578 | -3.008 | -1.155 | 1.333 | -1.172 | -0.381 | -0.775 | -0.693 | -2.175 | -1.307 | -0.679 |
| 04 | 2.318 | -1.965 | -0.094 | -0.828 | -0.818 | -3.186 | -2.531 | -2.526 | 0.198 | -2.642 | -0.979 | -3.181 | -0.871 | -3.004 | -0.858 | 0.11 |
| 05 | 2.256 | -1.828 | -0.382 | 0.611 | -1.331 | -4.367 | -4.367 | -4.367 | 0.038 | -3.078 | -1.126 | -0.966 | -1.217 | -3.2 | -3.191 | 0.26 |
| 06 | 0.392 | -0.704 | 2.197 | -1.742 | -1.809 | -3.149 | -3.197 | -4.367 | -1.892 | -4.367 | -0.197 | -2.571 | -0.855 | -2.275 | 1.087 | -3.201 |
| 07 | -0.282 | -2.628 | 0.322 | -4.367 | -0.588 | -4.367 | -2.626 | -4.367 | 2.141 | 0.122 | 1.135 | -2.571 | -2.572 | -4.367 | -1.593 | -4.367 |
| 08 | -1.26 | -3.174 | 2.257 | -4.367 | 0.024 | -4.367 | -1.727 | -4.367 | -1.19 | -4.367 | 1.49 | -4.367 | -4.367 | -4.367 | -2.571 | -4.367 |
| 09 | -4.367 | -4.367 | 0.462 | -4.367 | -4.367 | -4.367 | -3.174 | -4.367 | -4.367 | -4.367 | 2.654 | -4.367 | -4.367 | -4.367 | -4.367 | -4.367 |
| 10 | -4.367 | -4.367 | -4.367 | -4.367 | -4.367 | -4.367 | -4.367 | -4.367 | 2.761 | -4.367 | -4.367 | -4.367 | -4.367 | -4.367 | -4.367 | -4.367 |
| 11 | 2.752 | -4.367 | -4.367 | -1.913 | -4.367 | -4.367 | -4.367 | -4.367 | -4.367 | -4.367 | -4.367 | -4.367 | -4.367 | -4.367 | -4.367 | -4.367 |
| 12 | -1.234 | 1.53 | 2.379 | -4.367 | -4.367 | -4.367 | -4.367 | -4.367 | -4.367 | -4.367 | -4.367 | -4.367 | -3.088 | -4.367 | -2.166 | -4.367 |
| 13 | -4.367 | -3.171 | -4.367 | -1.222 | -1.335 | -1.478 | -4.367 | 1.423 | -2.219 | -2.265 | -1.544 | 2.352 | -4.367 | -4.367 | -4.367 | -4.367 |
| 14 | -4.367 | -4.367 | -1.024 | -4.367 | -3.191 | -4.367 | -1.14 | -4.367 | -4.367 | -4.367 | -1.544 | -4.367 | -3.058 | -0.682 | 2.667 | -4.367 |
| 15 | -3.058 | -4.367 | -3.191 | -4.367 | -1.336 | -2.482 | -2.432 | -2.213 | 2.326 | 0.154 | 1.26 | -1.007 | -4.367 | -4.367 | -4.367 | -4.367 |
| 16 | 1.86 | -1.003 | 1.18 | -0.661 | -0.925 | -2.196 | -1.933 | -0.474 | -0.191 | -0.27 | 0.484 | -0.788 | -1.992 | -3.162 | -1.272 | -3.19 |
| 17 | 1.845 | -1.134 | -0.111 | -1.438 | -0.815 | -0.931 | -3.049 | -0.917 | 1.174 | -0.507 | -0.326 | -0.299 | -1.351 | -0.705 | -0.852 | -0.787 |
| 18 | 0.805 | 1.301 | 1.177 | 0.103 | -0.445 | -0.44 | -2.174 | -0.883 | -0.114 | -0.552 | -0.824 | -1.697 | -0.843 | -0.772 | -1.12 | -0.487 |
| 19 | 0.467 | -0.188 | 0.025 | -0.285 | 0.408 | 0.222 | -1.394 | 0.857 | 0.259 | -0.355 | -0.149 | 0.235 | -1.318 | -0.547 | -0.764 | 0.008 |