Model info
| Transcription factor | SOX2 | ||||||||
| Model | SOX2_HUMAN.H10DI.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 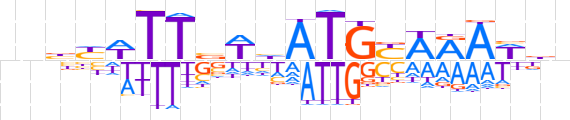 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 20 | ||||||||
| Quality | B | ||||||||
| Consensus | nnbYWTTbWhATGYWRAWdn | ||||||||
| wAUC | 0.9162770503705703 | ||||||||
| Best AUC | 0.9162770503705703 | ||||||||
| Benchmark datasets | 1 | ||||||||
| Aligned words | 421 | ||||||||
| TF family | SOX-related factors{4.1.1} | ||||||||
| TF subfamily | Group B{4.1.1.2} | ||||||||
| HGNC | 11195 | ||||||||
| EntrezGene | 6657 | ||||||||
| UniProt ID | SOX2_HUMAN | ||||||||
| UniProt AC | P48431 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 27.894 | 28.164 | 33.51 | 24.018 | 11.459 | 15.825 | 6.806 | 45.09 | 9.607 | 24.963 | 36.788 | 19.954 | 8.028 | 24.305 | 51.384 | 54.205 |
| 02 | 5.317 | 22.712 | 21.346 | 7.613 | 8.283 | 41.774 | 2.055 | 41.145 | 9.715 | 61.591 | 25.91 | 31.272 | 10.192 | 56.576 | 24.405 | 52.095 |
| 03 | 3.03 | 21.43 | 2.989 | 6.057 | 12.257 | 90.191 | 3.107 | 77.098 | 7.24 | 40.191 | 6.273 | 20.011 | 11.6 | 62.015 | 11.681 | 46.828 |
| 04 | 19.736 | 1.996 | 0.0 | 12.395 | 103.266 | 6.84 | 0.991 | 102.731 | 6.735 | 0.945 | 0.974 | 15.397 | 59.086 | 9.155 | 4.858 | 76.895 |
| 05 | 3.032 | 4.857 | 5.98 | 174.954 | 0.0 | 0.945 | 0.0 | 17.991 | 0.0 | 0.0 | 0.0 | 6.823 | 0.964 | 3.814 | 0.0 | 202.639 |
| 06 | 2.898 | 0.0 | 0.0 | 1.097 | 1.877 | 0.0 | 0.0 | 7.739 | 0.0 | 0.0 | 0.0 | 5.98 | 30.648 | 0.0 | 1.889 | 369.87 |
| 07 | 1.938 | 17.246 | 11.309 | 4.931 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.889 | 0.0 | 0.0 | 42.99 | 108.361 | 190.678 | 42.658 |
| 08 | 8.764 | 0.924 | 0.955 | 34.286 | 81.883 | 0.0 | 0.983 | 44.63 | 45.08 | 4.832 | 4.137 | 147.937 | 18.361 | 1.016 | 1.879 | 26.332 |
| 09 | 60.211 | 23.431 | 21.273 | 49.174 | 0.913 | 0.924 | 0.0 | 4.935 | 4.929 | 1.126 | 0.0 | 1.899 | 30.198 | 64.428 | 18.5 | 140.06 |
| 10 | 93.27 | 0.0 | 1.016 | 1.964 | 79.662 | 1.858 | 0.0 | 8.388 | 32.952 | 0.0 | 0.955 | 5.866 | 168.454 | 4.037 | 5.099 | 18.479 |
| 11 | 13.464 | 1.963 | 4.987 | 353.925 | 0.0 | 0.0 | 0.0 | 5.895 | 0.0 | 0.955 | 0.0 | 6.115 | 0.0 | 0.0 | 0.0 | 34.696 |
| 12 | 2.851 | 0.0 | 10.614 | 0.0 | 1.909 | 0.0 | 0.0 | 1.008 | 0.0 | 0.0 | 2.971 | 2.016 | 2.153 | 1.996 | 346.484 | 49.998 |
| 13 | 0.0 | 6.913 | 0.0 | 0.0 | 0.0 | 0.924 | 0.0 | 1.073 | 15.88 | 227.295 | 35.467 | 81.426 | 5.053 | 32.208 | 0.983 | 14.778 |
| 14 | 9.006 | 2.103 | 0.0 | 9.825 | 201.556 | 9.659 | 0.0 | 56.125 | 25.669 | 0.0 | 0.934 | 9.846 | 67.516 | 0.945 | 0.0 | 28.816 |
| 15 | 220.373 | 2.14 | 55.516 | 25.719 | 10.809 | 0.0 | 0.0 | 1.897 | 0.934 | 0.0 | 0.0 | 0.0 | 79.271 | 2.881 | 12.745 | 9.714 |
| 16 | 280.717 | 5.729 | 11.986 | 12.955 | 3.123 | 0.0 | 0.0 | 1.899 | 63.395 | 1.889 | 1.079 | 1.897 | 24.665 | 1.847 | 3.811 | 7.007 |
| 17 | 61.551 | 24.803 | 27.288 | 258.259 | 3.82 | 1.889 | 0.0 | 3.757 | 6.006 | 3.869 | 2.07 | 4.929 | 1.12 | 4.975 | 0.934 | 16.728 |
| 18 | 21.06 | 12.163 | 14.375 | 24.899 | 11.26 | 7.731 | 0.0 | 16.544 | 2.933 | 4.207 | 8.154 | 14.999 | 48.776 | 23.815 | 84.693 | 126.39 |
| 19 | 16.476 | 24.681 | 20.865 | 22.008 | 17.052 | 9.179 | 3.034 | 18.651 | 34.807 | 17.375 | 29.842 | 25.197 | 34.761 | 26.941 | 43.613 | 77.518 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.055 | 0.065 | 0.236 | -0.092 | -0.815 | -0.501 | -1.315 | 0.53 | -0.986 | -0.054 | 0.329 | -0.274 | -1.158 | -0.081 | 0.66 | 0.713 |
| 02 | -1.547 | -0.147 | -0.208 | -1.208 | -1.128 | 0.455 | -2.398 | 0.44 | -0.975 | 0.84 | -0.018 | 0.168 | -0.929 | 0.756 | -0.076 | 0.674 |
| 03 | -2.06 | -0.204 | -2.073 | -1.425 | -0.75 | 1.219 | -2.038 | 1.063 | -1.256 | 0.416 | -1.392 | -0.272 | -0.804 | 0.847 | -0.797 | 0.568 |
| 04 | -0.285 | -2.422 | -4.26 | -0.739 | 1.354 | -1.31 | -2.973 | 1.349 | -1.325 | -3.007 | -2.986 | -0.528 | 0.799 | -1.032 | -1.631 | 1.061 |
| 05 | -2.06 | -1.631 | -1.437 | 1.88 | -4.26 | -3.007 | -4.26 | -0.376 | -4.26 | -4.26 | -4.26 | -1.312 | -2.992 | -1.853 | -4.26 | 2.027 |
| 06 | -2.1 | -4.26 | -4.26 | -2.898 | -2.474 | -4.26 | -4.26 | -1.193 | -4.26 | -4.26 | -4.26 | -1.437 | 0.148 | -4.26 | -2.468 | 2.628 |
| 07 | -2.447 | -0.417 | -0.828 | -1.617 | -4.26 | -4.26 | -4.26 | -4.26 | -4.26 | -2.468 | -4.26 | -4.26 | 0.483 | 1.402 | 1.966 | 0.475 |
| 08 | -1.074 | -3.023 | -3.0 | 0.259 | 1.123 | -4.26 | -2.979 | 0.52 | 0.53 | -1.636 | -1.779 | 1.713 | -0.356 | -2.955 | -2.473 | -0.002 |
| 09 | 0.817 | -0.117 | -0.212 | 0.616 | -3.032 | -3.023 | -4.26 | -1.617 | -1.618 | -2.879 | -4.26 | -2.464 | 0.134 | 0.885 | -0.349 | 1.658 |
| 10 | 1.253 | -4.26 | -2.955 | -2.436 | 1.096 | -2.482 | -4.26 | -1.116 | 0.22 | -4.26 | -3.0 | -1.455 | 1.842 | -1.802 | -1.586 | -0.35 |
| 11 | -0.659 | -2.436 | -1.607 | 2.584 | -4.26 | -4.26 | -4.26 | -1.45 | -4.26 | -3.0 | -4.26 | -1.416 | -4.26 | -4.26 | -4.26 | 0.271 |
| 12 | -2.115 | -4.26 | -0.89 | -4.26 | -2.459 | -4.26 | -4.26 | -2.96 | -4.26 | -4.26 | -2.078 | -2.414 | -2.358 | -2.422 | 2.562 | 0.633 |
| 13 | -4.26 | -1.3 | -4.26 | -4.26 | -4.26 | -3.023 | -4.26 | -2.915 | -0.498 | 2.141 | 0.293 | 1.118 | -1.594 | 0.197 | -2.979 | -0.568 |
| 14 | -1.048 | -2.378 | -4.26 | -0.964 | 2.021 | -0.98 | -4.26 | 0.748 | -0.027 | -4.26 | -3.015 | -0.962 | 0.931 | -3.007 | -4.26 | 0.087 |
| 15 | 2.11 | -2.363 | 0.737 | -0.025 | -0.872 | -4.26 | -4.26 | -2.465 | -3.015 | -4.26 | -4.26 | -4.26 | 1.091 | -2.105 | -0.712 | -0.975 |
| 16 | 2.352 | -1.477 | -0.772 | -0.696 | -2.034 | -4.26 | -4.26 | -2.464 | 0.869 | -2.468 | -2.91 | -2.465 | -0.066 | -2.487 | -1.854 | -1.287 |
| 17 | 0.839 | -0.061 | 0.034 | 2.269 | -1.852 | -2.468 | -4.26 | -1.867 | -1.433 | -1.84 | -2.391 | -1.618 | -2.883 | -1.609 | -3.015 | -0.447 |
| 18 | -0.221 | -0.758 | -0.595 | -0.057 | -0.832 | -1.194 | -4.26 | -0.458 | -2.09 | -1.764 | -1.143 | -0.554 | 0.608 | -0.101 | 1.157 | 1.556 |
| 19 | -0.462 | -0.065 | -0.231 | -0.178 | -0.428 | -1.029 | -2.059 | -0.341 | 0.274 | -0.41 | 0.122 | -0.045 | 0.273 | 0.021 | 0.497 | 1.069 |