Model info
| Transcription factor | Prdm14 | ||||||||
| Model | PRD14_MOUSE.H10DI.C | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 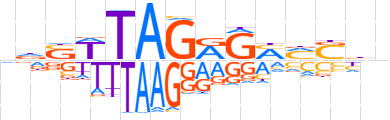 | ||||||||
| LOGO (reverse complement) | 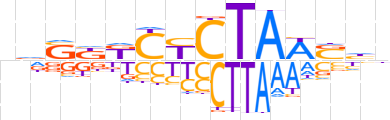 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 14 | ||||||||
| Quality | C | ||||||||
| Consensus | ndSWTAGRGMMShn | ||||||||
| wAUC | 0.8016261451947589 | ||||||||
| Best AUC | 0.8016261451947589 | ||||||||
| Benchmark datasets | 1 | ||||||||
| Aligned words | 511 | ||||||||
| TF family | More than 3 adjacent zinc finger factors{2.3.3} | ||||||||
| TF subfamily | unclassified{2.3.3.0} | ||||||||
| MGI | 3588194 | ||||||||
| EntrezGene | 383491 | ||||||||
| UniProt ID | PRD14_MOUSE | ||||||||
| UniProt AC | E9Q3T6 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 38.29 | 9.008 | 97.974 | 4.919 | 50.051 | 7.165 | 8.788 | 30.375 | 49.724 | 9.292 | 49.008 | 31.479 | 35.993 | 12.085 | 45.138 | 20.711 |
| 02 | 4.962 | 24.436 | 136.883 | 7.778 | 4.998 | 19.25 | 3.921 | 9.381 | 20.586 | 47.994 | 118.487 | 13.84 | 5.997 | 23.406 | 52.221 | 5.859 |
| 03 | 13.159 | 1.887 | 2.782 | 18.716 | 8.668 | 0.911 | 0.942 | 104.565 | 67.99 | 16.566 | 25.092 | 201.863 | 0.909 | 1.037 | 0.0 | 34.913 |
| 04 | 7.716 | 1.08 | 1.854 | 80.074 | 0.92 | 0.0 | 0.0 | 19.482 | 0.0 | 0.904 | 2.117 | 25.796 | 1.982 | 2.215 | 4.184 | 351.676 |
| 05 | 10.618 | 0.0 | 0.0 | 0.0 | 4.2 | 0.0 | 0.0 | 0.0 | 8.154 | 0.0 | 0.0 | 0.0 | 470.825 | 0.0 | 5.257 | 0.945 |
| 06 | 25.834 | 20.988 | 445.916 | 1.059 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 5.257 | 0.0 | 0.0 | 0.0 | 0.945 | 0.0 |
| 07 | 17.549 | 0.0 | 8.285 | 0.0 | 20.988 | 0.0 | 0.0 | 0.0 | 247.569 | 6.993 | 179.256 | 18.301 | 1.059 | 0.0 | 0.0 | 0.0 |
| 08 | 16.574 | 5.046 | 249.848 | 15.697 | 0.964 | 0.955 | 5.074 | 0.0 | 38.841 | 7.884 | 134.128 | 6.687 | 0.0 | 0.0 | 16.317 | 1.983 |
| 09 | 41.353 | 7.001 | 0.0 | 8.025 | 7.858 | 2.873 | 0.0 | 3.154 | 241.208 | 101.2 | 11.924 | 51.036 | 12.128 | 7.59 | 0.977 | 3.673 |
| 10 | 69.211 | 171.84 | 38.943 | 22.551 | 31.29 | 77.552 | 0.948 | 8.875 | 4.703 | 7.194 | 1.004 | 0.0 | 25.664 | 30.265 | 3.091 | 6.868 |
| 11 | 3.181 | 67.49 | 43.586 | 16.611 | 12.21 | 231.282 | 10.997 | 32.363 | 5.099 | 21.999 | 10.66 | 6.23 | 0.946 | 21.472 | 13.925 | 1.951 |
| 12 | 7.819 | 2.795 | 9.826 | 0.995 | 64.75 | 129.854 | 10.485 | 137.153 | 3.973 | 29.129 | 30.016 | 16.05 | 9.58 | 16.873 | 11.202 | 19.5 |
| 13 | 21.195 | 13.479 | 44.173 | 7.275 | 68.536 | 24.963 | 16.045 | 69.107 | 18.189 | 13.198 | 21.074 | 9.067 | 24.091 | 41.325 | 69.922 | 38.36 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.201 | -1.214 | 1.134 | -1.785 | 0.466 | -1.432 | -1.238 | -0.028 | 0.46 | -1.184 | 0.446 | 0.007 | 0.14 | -0.931 | 0.364 | -0.405 |
| 02 | -1.777 | -0.243 | 1.468 | -1.354 | -1.77 | -0.477 | -1.994 | -1.175 | -0.411 | 0.425 | 1.324 | -0.799 | -1.6 | -0.285 | 0.509 | -1.622 |
| 03 | -0.848 | -2.632 | -2.3 | -0.504 | -1.251 | -3.192 | -3.169 | 1.199 | 0.771 | -0.624 | -0.216 | 1.855 | -3.194 | -3.1 | -4.4 | 0.11 |
| 04 | -1.362 | -3.07 | -2.647 | 0.933 | -3.186 | -4.4 | -4.4 | -0.465 | -4.4 | -3.198 | -2.536 | -0.189 | -2.591 | -2.497 | -1.934 | 2.409 |
| 05 | -1.056 | -4.4 | -4.4 | -4.4 | -1.931 | -4.4 | -4.4 | -4.4 | -1.309 | -4.4 | -4.4 | -4.4 | 2.701 | -4.4 | -1.723 | -3.166 |
| 06 | -0.188 | -0.392 | 2.647 | -3.084 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -1.723 | -4.4 | -4.4 | -4.4 | -3.166 | -4.4 |
| 07 | -0.567 | -4.4 | -1.294 | -4.4 | -0.392 | -4.4 | -4.4 | -4.4 | 2.059 | -1.455 | 1.737 | -0.526 | -3.084 | -4.4 | -4.4 | -4.4 |
| 08 | -0.623 | -1.762 | 2.068 | -0.676 | -3.152 | -3.159 | -1.756 | -4.4 | 0.215 | -1.341 | 1.447 | -1.498 | -4.4 | -4.4 | -0.639 | -2.591 |
| 09 | 0.277 | -1.454 | -4.4 | -1.325 | -1.345 | -2.272 | -4.4 | -2.189 | 2.033 | 1.167 | -0.944 | 0.486 | -0.927 | -1.378 | -3.143 | -2.053 |
| 10 | 0.788 | 1.694 | 0.218 | -0.321 | 0.001 | 0.902 | -3.164 | -1.228 | -1.827 | -1.429 | -3.123 | -4.4 | -0.194 | -0.032 | -2.207 | -1.473 |
| 11 | -2.182 | 0.763 | 0.329 | -0.621 | -0.921 | 1.991 | -1.022 | 0.035 | -1.752 | -0.346 | -1.052 | -1.565 | -3.166 | -0.37 | -0.793 | -2.604 |
| 12 | -1.349 | -2.296 | -1.131 | -3.13 | 0.722 | 1.415 | -1.068 | 1.47 | -1.981 | -0.069 | -0.04 | -0.655 | -1.155 | -0.606 | -1.004 | -0.464 |
| 13 | -0.382 | -0.825 | 0.343 | -1.418 | 0.779 | -0.222 | -0.655 | 0.787 | -0.532 | -0.845 | -0.388 | -1.208 | -0.257 | 0.276 | 0.799 | 0.203 |