Model info
| Transcription factor | PPARG | ||||||||
| Model | PPARG_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 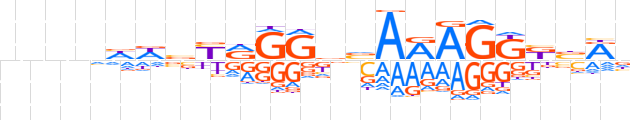 | ||||||||
| LOGO (reverse complement) | 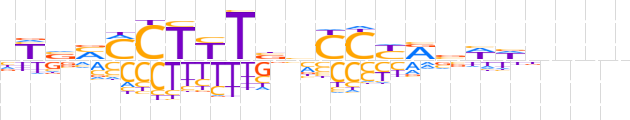 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 22 | ||||||||
| Quality | A | ||||||||
| Consensus | nnnnhWbYRGGdvARAGKKbRn | ||||||||
| wAUC | 0.8695454840686423 | ||||||||
| Best AUC | 0.8816948819498693 | ||||||||
| Benchmark datasets | 4 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | Thyroid hormone receptor-related factors (NR1){2.1.2} | ||||||||
| TF subfamily | PPAR (NR1C){2.1.2.5} | ||||||||
| HGNC | 9236 | ||||||||
| EntrezGene | 5468 | ||||||||
| UniProt ID | PPARG_HUMAN | ||||||||
| UniProt AC | P37231 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 38.715 | 18.765 | 62.951 | 20.491 | 47.784 | 12.213 | 6.501 | 34.191 | 39.143 | 29.375 | 57.689 | 19.019 | 7.452 | 28.809 | 44.611 | 32.29 |
| 02 | 30.534 | 20.608 | 66.598 | 15.354 | 28.883 | 28.662 | 3.021 | 28.595 | 33.768 | 58.871 | 56.894 | 22.221 | 13.476 | 23.589 | 37.158 | 31.768 |
| 03 | 50.078 | 13.379 | 24.413 | 18.79 | 27.82 | 70.643 | 3.123 | 30.144 | 58.152 | 21.718 | 57.596 | 26.205 | 16.552 | 17.795 | 28.931 | 34.659 |
| 04 | 109.59 | 9.352 | 20.607 | 13.053 | 74.199 | 17.197 | 2.124 | 30.015 | 67.416 | 18.923 | 19.374 | 8.352 | 32.926 | 18.647 | 20.486 | 37.739 |
| 05 | 164.725 | 2.66 | 41.23 | 75.515 | 37.992 | 3.568 | 2.502 | 20.056 | 40.288 | 1.691 | 9.62 | 10.992 | 24.591 | 9.808 | 10.016 | 44.743 |
| 06 | 17.594 | 86.65 | 114.078 | 49.274 | 2.992 | 5.494 | 0.899 | 8.342 | 15.228 | 21.924 | 19.803 | 6.414 | 14.958 | 57.661 | 45.737 | 32.951 |
| 07 | 6.786 | 3.971 | 9.34 | 30.675 | 7.297 | 28.848 | 6.685 | 128.9 | 10.008 | 27.817 | 22.421 | 120.272 | 0.854 | 15.031 | 28.1 | 52.996 |
| 08 | 15.226 | 0.0 | 9.719 | 0.0 | 49.248 | 1.954 | 11.198 | 13.267 | 37.786 | 5.04 | 15.652 | 8.069 | 98.405 | 10.49 | 210.951 | 12.996 |
| 09 | 24.988 | 0.0 | 168.782 | 6.894 | 7.853 | 0.0 | 7.217 | 2.414 | 18.139 | 0.0 | 199.769 | 29.612 | 1.076 | 0.0 | 27.649 | 5.607 |
| 10 | 3.843 | 1.089 | 44.345 | 2.779 | 0.0 | 0.0 | 0.0 | 0.0 | 75.012 | 3.708 | 306.55 | 18.146 | 3.93 | 1.12 | 37.771 | 1.706 |
| 11 | 19.737 | 5.713 | 52.638 | 4.698 | 4.081 | 0.0 | 0.0 | 1.836 | 55.982 | 66.231 | 142.045 | 124.408 | 3.667 | 3.502 | 10.424 | 5.038 |
| 12 | 6.826 | 47.489 | 18.331 | 10.821 | 14.078 | 44.182 | 1.943 | 15.243 | 28.086 | 107.063 | 55.235 | 14.723 | 18.427 | 46.848 | 55.573 | 15.131 |
| 13 | 61.563 | 0.0 | 5.855 | 0.0 | 235.276 | 1.932 | 4.261 | 4.113 | 126.326 | 0.0 | 4.757 | 0.0 | 51.561 | 1.117 | 2.366 | 0.874 |
| 14 | 321.354 | 14.655 | 123.337 | 15.38 | 3.049 | 0.0 | 0.0 | 0.0 | 12.319 | 3.215 | 0.854 | 0.85 | 1.904 | 0.0 | 2.187 | 0.896 |
| 15 | 279.436 | 0.0 | 57.312 | 1.877 | 16.815 | 1.056 | 0.0 | 0.0 | 105.206 | 0.878 | 20.294 | 0.0 | 13.362 | 0.0 | 2.869 | 0.896 |
| 16 | 49.0 | 1.97 | 361.259 | 2.59 | 0.0 | 0.0 | 1.056 | 0.878 | 15.364 | 0.0 | 61.403 | 3.708 | 0.0 | 0.0 | 2.772 | 0.0 |
| 17 | 4.414 | 0.0 | 58.851 | 1.099 | 0.0 | 0.0 | 1.97 | 0.0 | 46.751 | 4.419 | 306.403 | 68.917 | 0.0 | 0.0 | 7.176 | 0.0 |
| 18 | 1.35 | 3.65 | 31.002 | 15.163 | 1.189 | 1.051 | 1.046 | 1.133 | 12.309 | 51.901 | 127.441 | 182.749 | 1.322 | 3.677 | 17.96 | 47.056 |
| 19 | 2.199 | 4.379 | 9.593 | 0.0 | 13.061 | 31.827 | 2.169 | 13.222 | 23.594 | 91.202 | 35.724 | 26.929 | 14.77 | 142.781 | 48.695 | 39.856 |
| 20 | 34.838 | 5.433 | 10.441 | 2.912 | 212.676 | 13.298 | 11.472 | 32.744 | 74.033 | 10.868 | 10.198 | 1.082 | 36.299 | 6.689 | 24.457 | 12.562 |
| 21 | 88.681 | 73.607 | 114.748 | 80.809 | 12.643 | 11.705 | 1.646 | 10.293 | 11.615 | 11.695 | 17.432 | 15.826 | 2.583 | 14.141 | 19.152 | 13.424 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.212 | -0.502 | 0.694 | -0.416 | 0.42 | -0.921 | -1.524 | 0.089 | 0.223 | -0.061 | 0.607 | -0.489 | -1.395 | -0.08 | 0.352 | 0.032 |
| 02 | -0.023 | -0.41 | 0.75 | -0.698 | -0.078 | -0.085 | -2.228 | -0.088 | 0.077 | 0.628 | 0.594 | -0.336 | -0.825 | -0.277 | 0.171 | 0.016 |
| 03 | 0.467 | -0.832 | -0.243 | -0.501 | -0.115 | 0.809 | -2.198 | -0.036 | 0.615 | -0.358 | 0.606 | -0.174 | -0.625 | -0.554 | -0.076 | 0.102 |
| 04 | 1.246 | -1.178 | -0.41 | -0.856 | 0.858 | -0.587 | -2.533 | -0.04 | 0.762 | -0.494 | -0.471 | -1.286 | 0.052 | -0.508 | -0.416 | 0.187 |
| 05 | 1.652 | -2.34 | 0.274 | 0.875 | 0.193 | -2.079 | -2.393 | -0.437 | 0.251 | -2.722 | -1.151 | -1.022 | -0.236 | -1.132 | -1.112 | 0.355 |
| 06 | -0.565 | 1.012 | 1.286 | 0.451 | -2.236 | -1.682 | -3.202 | -1.288 | -0.706 | -0.349 | -0.449 | -1.537 | -0.723 | 0.607 | 0.377 | 0.052 |
| 07 | -1.484 | -1.982 | -1.179 | -0.018 | -1.415 | -0.079 | -1.498 | 1.408 | -1.113 | -0.115 | -0.327 | 1.339 | -3.237 | -0.719 | -0.105 | 0.523 |
| 08 | -0.706 | -4.4 | -1.141 | -4.4 | 0.45 | -2.603 | -1.005 | -0.84 | 0.188 | -1.763 | -0.679 | -1.319 | 1.139 | -1.068 | 1.899 | -0.86 |
| 09 | -0.221 | -4.4 | 1.677 | -1.469 | -1.345 | -4.4 | -1.426 | -2.424 | -0.535 | -4.4 | 1.845 | -0.053 | -3.073 | -4.4 | -0.121 | -1.663 |
| 10 | -2.012 | -3.064 | 0.346 | -2.302 | -4.4 | -4.4 | -4.4 | -4.4 | 0.868 | -2.044 | 2.272 | -0.535 | -1.992 | -3.043 | 0.187 | -2.715 |
| 11 | -0.452 | -1.646 | 0.516 | -1.828 | -1.957 | -4.4 | -4.4 | -2.655 | 0.578 | 0.745 | 1.505 | 1.372 | -2.054 | -2.096 | -1.074 | -1.763 |
| 12 | -1.478 | 0.414 | -0.525 | -1.038 | -0.783 | 0.343 | -2.608 | -0.705 | -0.105 | 1.223 | 0.564 | -0.739 | -0.52 | 0.401 | 0.57 | -0.712 |
| 13 | 0.672 | -4.4 | -1.623 | -4.4 | 2.008 | -2.613 | -1.918 | -1.95 | 1.388 | -4.4 | -1.816 | -4.4 | 0.496 | -3.046 | -2.441 | -3.221 |
| 14 | 2.319 | -0.743 | 1.364 | -0.696 | -2.22 | -4.4 | -4.4 | -4.4 | -0.912 | -2.172 | -3.237 | -3.24 | -2.625 | -4.4 | -2.508 | -3.204 |
| 15 | 2.18 | -4.4 | 0.601 | -2.637 | -0.609 | -3.087 | -4.4 | -4.4 | 1.205 | -3.218 | -0.425 | -4.4 | -0.833 | -4.4 | -2.273 | -3.204 |
| 16 | 0.445 | -2.596 | 2.436 | -2.363 | -4.4 | -4.4 | -3.087 | -3.218 | -0.697 | -4.4 | 0.669 | -2.044 | -4.4 | -4.4 | -2.304 | -4.4 |
| 17 | -1.885 | -4.4 | 0.627 | -3.058 | -4.4 | -4.4 | -2.596 | -4.4 | 0.399 | -1.884 | 2.272 | 0.784 | -4.4 | -4.4 | -1.431 | -4.4 |
| 18 | -2.902 | -2.059 | -0.008 | -0.71 | -2.998 | -3.09 | -3.094 | -3.035 | -0.913 | 0.502 | 1.396 | 1.756 | -2.917 | -2.052 | -0.545 | 0.405 |
| 19 | -2.504 | -1.893 | -1.154 | -4.4 | -0.855 | 0.018 | -2.516 | -0.844 | -0.277 | 1.063 | 0.132 | -0.147 | -0.736 | 1.51 | 0.439 | 0.241 |
| 20 | 0.107 | -1.693 | -1.072 | -2.26 | 1.907 | -0.838 | -0.981 | 0.046 | 0.855 | -1.033 | -1.095 | -3.069 | 0.148 | -1.497 | -0.242 | -0.893 |
| 21 | 1.035 | 0.85 | 1.292 | 0.943 | -0.887 | -0.962 | -2.744 | -1.086 | -0.969 | -0.963 | -0.574 | -0.668 | -2.365 | -0.778 | -0.482 | -0.829 |