Model info
| Transcription factor | Myc | ||||||||
| Model | MYC_MOUSE.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 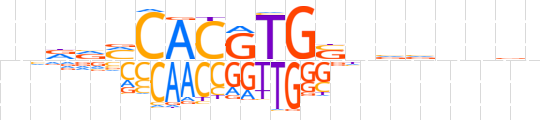 | ||||||||
| LOGO (reverse complement) | 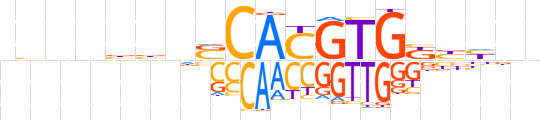 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 19 | ||||||||
| Quality | A | ||||||||
| Consensus | nndvMCACGTGSnvvnnvn | ||||||||
| wAUC | 0.8562701197561228 | ||||||||
| Best AUC | 0.8929962114698462 | ||||||||
| Benchmark datasets | 3 | ||||||||
| Aligned words | 501 | ||||||||
| TF family | bHLH-ZIP factors{1.2.6} | ||||||||
| TF subfamily | Myc / Max factors{1.2.6.5} | ||||||||
| MGI | 97250 | ||||||||
| EntrezGene | 17869 | ||||||||
| UniProt ID | MYC_MOUSE | ||||||||
| UniProt AC | P01108 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 50.793 | 32.692 | 37.254 | 7.893 | 44.828 | 28.766 | 10.65 | 29.188 | 45.189 | 74.778 | 35.404 | 19.668 | 17.919 | 34.736 | 14.412 | 15.831 |
| 02 | 55.564 | 10.951 | 78.699 | 13.515 | 102.994 | 9.644 | 29.753 | 28.582 | 55.795 | 3.997 | 32.906 | 5.022 | 18.178 | 4.054 | 41.571 | 8.775 |
| 03 | 77.881 | 34.892 | 103.214 | 16.544 | 9.068 | 9.445 | 9.244 | 0.89 | 69.023 | 16.186 | 90.82 | 6.898 | 7.05 | 16.026 | 30.706 | 2.112 |
| 04 | 29.923 | 81.984 | 48.293 | 2.822 | 23.966 | 45.389 | 6.048 | 1.147 | 44.076 | 153.417 | 30.721 | 5.771 | 8.128 | 9.657 | 8.659 | 0.0 |
| 05 | 3.174 | 102.92 | 0.0 | 0.0 | 18.629 | 269.666 | 2.152 | 0.0 | 3.921 | 89.799 | 0.0 | 0.0 | 0.0 | 8.778 | 0.962 | 0.0 |
| 06 | 16.935 | 0.0 | 8.789 | 0.0 | 431.13 | 1.898 | 27.45 | 10.684 | 2.119 | 0.0 | 0.994 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 07 | 1.998 | 401.952 | 4.869 | 41.366 | 0.0 | 1.898 | 0.0 | 0.0 | 1.091 | 32.062 | 0.989 | 3.091 | 0.0 | 8.81 | 0.0 | 1.874 |
| 08 | 3.089 | 0.0 | 0.0 | 0.0 | 105.301 | 8.592 | 330.829 | 0.0 | 2.081 | 0.0 | 3.777 | 0.0 | 0.893 | 0.0 | 45.438 | 0.0 |
| 09 | 3.823 | 0.0 | 0.0 | 107.541 | 0.0 | 0.0 | 0.0 | 8.592 | 11.471 | 17.278 | 0.993 | 350.301 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 0.929 | 0.0 | 14.365 | 0.0 | 1.005 | 0.0 | 13.308 | 2.965 | 0.0 | 0.0 | 0.993 | 0.0 | 0.938 | 1.928 | 458.347 | 5.221 |
| 11 | 0.0 | 2.873 | 0.0 | 0.0 | 0.863 | 0.0 | 0.0 | 1.065 | 20.112 | 140.088 | 283.595 | 43.219 | 0.0 | 3.984 | 2.131 | 2.071 |
| 12 | 2.06 | 7.664 | 8.984 | 2.267 | 39.994 | 25.308 | 27.032 | 54.61 | 69.758 | 66.385 | 48.194 | 101.388 | 11.625 | 6.404 | 17.456 | 10.871 |
| 13 | 37.729 | 16.626 | 55.455 | 13.625 | 49.32 | 20.38 | 28.881 | 7.18 | 19.394 | 36.005 | 30.682 | 15.586 | 30.263 | 31.318 | 94.996 | 12.56 |
| 14 | 41.706 | 31.122 | 56.788 | 7.09 | 38.242 | 26.564 | 27.646 | 11.878 | 46.879 | 59.075 | 85.806 | 18.253 | 9.956 | 18.865 | 19.157 | 0.973 |
| 15 | 40.012 | 35.128 | 44.937 | 16.707 | 31.927 | 39.905 | 20.535 | 43.258 | 28.991 | 95.449 | 50.98 | 13.977 | 4.915 | 19.273 | 11.027 | 2.98 |
| 16 | 27.354 | 19.801 | 45.831 | 12.858 | 37.489 | 37.714 | 34.233 | 80.32 | 31.559 | 31.638 | 31.202 | 33.08 | 9.126 | 13.69 | 42.812 | 11.294 |
| 17 | 22.672 | 23.381 | 53.606 | 5.87 | 39.402 | 28.243 | 23.434 | 11.764 | 45.029 | 25.218 | 59.793 | 24.038 | 14.052 | 62.249 | 49.321 | 11.929 |
| 18 | 24.326 | 34.382 | 54.436 | 8.013 | 69.635 | 24.511 | 8.679 | 36.265 | 47.811 | 53.841 | 58.913 | 25.588 | 8.058 | 14.86 | 21.996 | 8.686 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.481 | 0.045 | 0.174 | -1.34 | 0.357 | -0.082 | -1.053 | -0.067 | 0.365 | 0.865 | 0.123 | -0.456 | -0.547 | 0.105 | -0.76 | -0.668 |
| 02 | 0.57 | -1.026 | 0.916 | -0.822 | 1.184 | -1.149 | -0.048 | -0.088 | 0.574 | -1.976 | 0.051 | -1.766 | -0.533 | -1.963 | 0.282 | -1.239 |
| 03 | 0.906 | 0.109 | 1.186 | -0.625 | -1.208 | -1.169 | -1.189 | -3.209 | 0.786 | -0.646 | 1.059 | -1.468 | -1.448 | -0.656 | -0.017 | -2.538 |
| 04 | -0.043 | 0.957 | 0.431 | -2.288 | -0.262 | 0.369 | -1.592 | -3.026 | 0.34 | 1.581 | -0.017 | -1.636 | -1.312 | -1.147 | -1.252 | -4.4 |
| 05 | -2.184 | 1.183 | -4.4 | -4.4 | -0.509 | 2.144 | -2.522 | -4.4 | -1.994 | 1.048 | -4.4 | -4.4 | -4.4 | -1.239 | -3.154 | -4.4 |
| 06 | -0.602 | -4.4 | -1.238 | -4.4 | 2.613 | -2.627 | -0.128 | -1.05 | -2.535 | -4.4 | -3.13 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 |
| 07 | -2.585 | 2.543 | -1.795 | 0.277 | -4.4 | -2.627 | -4.4 | -4.4 | -3.062 | 0.025 | -3.134 | -2.208 | -4.4 | -1.235 | -4.4 | -2.638 |
| 08 | -2.208 | -4.4 | -4.4 | -4.4 | 1.206 | -1.259 | 2.348 | -4.4 | -2.55 | -4.4 | -2.027 | -4.4 | -3.207 | -4.4 | 0.37 | -4.4 |
| 09 | -2.017 | -4.4 | -4.4 | 1.227 | -4.4 | -4.4 | -4.4 | -1.259 | -0.981 | -0.583 | -3.131 | 2.406 | -4.4 | -4.4 | -4.4 | -4.4 |
| 10 | -3.179 | -4.4 | -0.763 | -4.4 | -3.122 | -4.4 | -0.837 | -2.244 | -4.4 | -4.4 | -3.131 | -4.4 | -3.172 | -2.614 | 2.674 | -1.73 |
| 11 | -4.4 | -2.272 | -4.4 | -4.4 | -3.23 | -4.4 | -4.4 | -3.08 | -0.434 | 1.491 | 2.195 | 0.321 | -4.4 | -1.979 | -2.53 | -2.554 |
| 12 | -2.559 | -1.368 | -1.217 | -2.478 | 0.244 | -0.208 | -0.143 | 0.553 | 0.796 | 0.747 | 0.429 | 1.168 | -0.968 | -1.539 | -0.573 | -1.033 |
| 13 | 0.186 | -0.62 | 0.568 | -0.814 | 0.452 | -0.421 | -0.078 | -1.43 | -0.47 | 0.14 | -0.018 | -0.683 | -0.032 | 0.002 | 1.104 | -0.893 |
| 14 | 0.286 | -0.004 | 0.592 | -1.442 | 0.2 | -0.16 | -0.121 | -0.948 | 0.401 | 0.631 | 1.002 | -0.529 | -1.118 | -0.497 | -0.482 | -3.146 |
| 15 | 0.244 | 0.116 | 0.359 | -0.616 | 0.021 | 0.242 | -0.413 | 0.322 | -0.074 | 1.108 | 0.485 | -0.79 | -1.786 | -0.476 | -1.019 | -2.24 |
| 16 | -0.131 | -0.449 | 0.379 | -0.871 | 0.18 | 0.186 | 0.09 | 0.936 | 0.01 | 0.012 | -0.002 | 0.056 | -1.202 | -0.81 | 0.311 | -0.996 |
| 17 | -0.316 | -0.286 | 0.535 | -1.62 | 0.229 | -0.1 | -0.284 | -0.957 | 0.362 | -0.212 | 0.643 | -0.259 | -0.784 | 0.683 | 0.452 | -0.943 |
| 18 | -0.247 | 0.094 | 0.55 | -1.326 | 0.794 | -0.24 | -1.25 | 0.147 | 0.421 | 0.539 | 0.628 | -0.197 | -1.321 | -0.73 | -0.346 | -1.249 |