Model info
| Transcription factor | Max | ||||||||
| Model | MAX_MOUSE.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 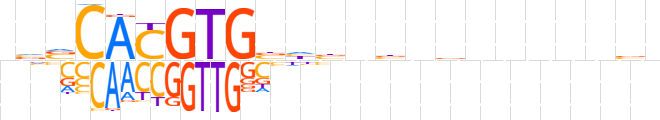 | ||||||||
| LOGO (reverse complement) | 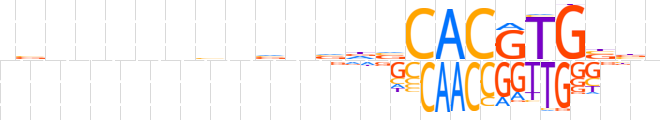 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Consensus | nvvCACGTGbhbnbnbnnnnnbn | ||||||||
| wAUC | 0.8306275995481456 | ||||||||
| Best AUC | 0.8968438650315729 | ||||||||
| Benchmark datasets | 4 | ||||||||
| Aligned words | 514 | ||||||||
| TF family | bHLH-ZIP factors{1.2.6} | ||||||||
| TF subfamily | Myc / Max factors{1.2.6.5} | ||||||||
| MGI | 96921 | ||||||||
| EntrezGene | |||||||||
| UniProt ID | MAX_MOUSE | ||||||||
| UniProt AC | P28574 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 38.365 | 41.11 | 50.425 | 10.558 | 49.538 | 34.587 | 33.663 | 12.399 | 32.591 | 57.905 | 39.053 | 3.911 | 18.436 | 29.009 | 41.104 | 7.344 |
| 02 | 27.782 | 78.154 | 31.12 | 1.875 | 40.295 | 71.05 | 47.683 | 3.584 | 22.058 | 83.902 | 39.256 | 19.029 | 6.144 | 16.819 | 11.249 | 0.0 |
| 03 | 3.721 | 92.558 | 0.0 | 0.0 | 4.977 | 243.99 | 0.957 | 0.0 | 0.85 | 128.458 | 0.0 | 0.0 | 0.0 | 23.662 | 0.826 | 0.0 |
| 04 | 5.729 | 1.794 | 2.024 | 0.0 | 454.068 | 0.893 | 21.249 | 12.459 | 0.0 | 0.826 | 0.957 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 05 | 0.0 | 363.236 | 1.858 | 94.704 | 0.0 | 0.0 | 0.893 | 2.62 | 0.0 | 23.371 | 0.0 | 0.86 | 0.0 | 11.217 | 0.0 | 1.242 |
| 06 | 0.0 | 0.0 | 0.0 | 0.0 | 4.441 | 5.492 | 384.992 | 2.9 | 0.0 | 1.858 | 0.893 | 0.0 | 0.824 | 0.838 | 97.764 | 0.0 |
| 07 | 0.0 | 0.0 | 0.0 | 5.264 | 0.0 | 0.0 | 0.0 | 8.188 | 5.602 | 9.238 | 2.636 | 466.172 | 0.0 | 0.0 | 0.0 | 2.9 |
| 08 | 0.0 | 0.0 | 5.602 | 0.0 | 0.0 | 1.833 | 4.63 | 2.775 | 0.838 | 0.0 | 1.798 | 0.0 | 3.752 | 0.853 | 472.381 | 5.538 |
| 09 | 0.0 | 1.855 | 2.735 | 0.0 | 0.0 | 2.686 | 0.0 | 0.0 | 59.011 | 255.29 | 99.739 | 70.371 | 0.0 | 1.825 | 2.744 | 3.744 |
| 10 | 6.709 | 46.383 | 1.116 | 4.802 | 58.784 | 42.842 | 23.167 | 136.863 | 20.032 | 39.196 | 4.296 | 41.695 | 12.714 | 22.032 | 0.988 | 38.381 |
| 11 | 7.014 | 43.202 | 33.886 | 14.137 | 32.886 | 75.7 | 14.186 | 27.681 | 1.05 | 17.013 | 7.365 | 4.138 | 19.594 | 100.328 | 48.804 | 53.015 |
| 12 | 17.976 | 16.68 | 25.888 | 0.0 | 66.675 | 71.989 | 45.54 | 52.041 | 20.945 | 36.156 | 30.981 | 16.158 | 7.603 | 29.327 | 35.761 | 26.281 |
| 13 | 15.662 | 33.119 | 43.503 | 20.914 | 14.374 | 57.366 | 30.639 | 51.772 | 7.135 | 70.435 | 36.774 | 23.828 | 7.49 | 40.64 | 24.865 | 21.484 |
| 14 | 14.159 | 8.592 | 14.36 | 7.55 | 54.055 | 66.366 | 26.175 | 54.963 | 21.074 | 44.158 | 40.988 | 29.56 | 9.402 | 23.694 | 53.685 | 31.216 |
| 15 | 11.426 | 18.476 | 49.861 | 18.929 | 34.121 | 51.526 | 28.04 | 29.125 | 21.669 | 44.887 | 39.315 | 29.336 | 2.967 | 31.434 | 67.24 | 21.65 |
| 16 | 10.315 | 12.103 | 40.076 | 7.687 | 40.476 | 32.303 | 35.031 | 38.513 | 39.417 | 60.933 | 51.188 | 32.917 | 11.991 | 13.876 | 55.74 | 17.434 |
| 17 | 10.0 | 34.021 | 44.418 | 13.76 | 13.869 | 41.471 | 29.149 | 34.726 | 36.315 | 61.197 | 50.334 | 34.188 | 10.285 | 26.966 | 39.453 | 19.847 |
| 18 | 19.048 | 14.109 | 22.071 | 15.241 | 47.482 | 54.678 | 31.096 | 30.398 | 36.091 | 58.02 | 40.85 | 28.393 | 17.922 | 36.055 | 32.066 | 16.48 |
| 19 | 32.278 | 26.766 | 48.293 | 13.206 | 45.416 | 57.929 | 10.887 | 48.63 | 27.964 | 46.772 | 32.344 | 19.002 | 14.586 | 33.416 | 22.662 | 19.849 |
| 20 | 16.543 | 37.428 | 48.189 | 18.084 | 32.622 | 57.43 | 31.651 | 43.18 | 15.365 | 43.446 | 34.818 | 20.558 | 11.973 | 27.186 | 35.353 | 26.175 |
| 21 | 12.182 | 25.907 | 28.101 | 10.312 | 21.865 | 79.047 | 32.793 | 31.786 | 16.976 | 74.614 | 30.677 | 27.745 | 10.918 | 35.56 | 41.299 | 20.219 |
| 22 | 15.432 | 17.428 | 20.663 | 8.419 | 60.795 | 66.684 | 27.512 | 60.136 | 21.125 | 62.617 | 32.896 | 16.232 | 4.965 | 29.135 | 36.313 | 19.65 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.203 | 0.271 | 0.474 | -1.061 | 0.456 | 0.1 | 0.073 | -0.906 | 0.042 | 0.611 | 0.22 | -1.996 | -0.519 | -0.073 | 0.271 | -1.409 |
| 02 | -0.116 | 0.909 | -0.004 | -2.638 | 0.251 | 0.814 | 0.418 | -2.075 | -0.343 | 0.98 | 0.226 | -0.488 | -1.578 | -0.609 | -1.0 | -4.4 |
| 03 | -2.041 | 1.078 | -4.4 | -4.4 | -1.774 | 2.044 | -3.157 | -4.4 | -3.24 | 1.404 | -4.4 | -4.4 | -4.4 | -0.274 | -3.26 | -4.4 |
| 04 | -1.643 | -2.674 | -2.574 | -4.4 | 2.665 | -3.207 | -0.38 | -0.901 | -4.4 | -3.26 | -3.157 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 |
| 05 | -4.4 | 2.442 | -2.645 | 1.1 | -4.4 | -4.4 | -3.207 | -2.353 | -4.4 | -0.286 | -4.4 | -3.233 | -4.4 | -1.003 | -4.4 | -2.966 |
| 06 | -4.4 | -4.4 | -4.4 | -4.4 | -1.88 | -1.683 | 2.5 | -2.264 | -4.4 | -2.645 | -3.207 | -4.4 | -3.262 | -3.25 | 1.132 | -4.4 |
| 07 | -4.4 | -4.4 | -4.4 | -1.722 | -4.4 | -4.4 | -4.4 | -1.305 | -1.664 | -1.19 | -2.348 | 2.691 | -4.4 | -4.4 | -4.4 | -2.264 |
| 08 | -4.4 | -4.4 | -1.664 | -4.4 | -4.4 | -2.656 | -1.841 | -2.303 | -3.25 | -4.4 | -2.672 | -4.4 | -2.034 | -3.238 | 2.704 | -1.675 |
| 09 | -4.4 | -2.646 | -2.315 | -4.4 | -4.4 | -2.331 | -4.4 | -4.4 | 0.63 | 2.09 | 1.152 | 0.805 | -4.4 | -2.66 | -2.312 | -2.036 |
| 10 | -1.495 | 0.391 | -3.046 | -1.808 | 0.626 | 0.312 | -0.295 | 1.467 | -0.438 | 0.224 | -1.91 | 0.285 | -0.882 | -0.344 | -3.135 | 0.203 |
| 11 | -1.453 | 0.32 | 0.08 | -0.778 | 0.05 | 0.878 | -0.775 | -0.12 | -3.091 | -0.598 | -1.406 | -1.944 | -0.46 | 1.158 | 0.441 | 0.523 |
| 12 | -0.544 | -0.617 | -0.186 | -4.4 | 0.751 | 0.828 | 0.373 | 0.505 | -0.394 | 0.144 | -0.009 | -0.648 | -1.376 | -0.063 | 0.133 | -0.171 |
| 13 | -0.679 | 0.057 | 0.327 | -0.396 | -0.762 | 0.602 | -0.019 | 0.5 | -1.436 | 0.806 | 0.161 | -0.267 | -1.39 | 0.26 | -0.225 | -0.369 |
| 14 | -0.777 | -1.259 | -0.763 | -1.383 | 0.543 | 0.747 | -0.175 | 0.559 | -0.388 | 0.342 | 0.268 | -0.055 | -1.173 | -0.273 | 0.536 | -0.001 |
| 15 | -0.985 | -0.517 | 0.463 | -0.493 | 0.087 | 0.495 | -0.107 | -0.07 | -0.361 | 0.358 | 0.227 | -0.062 | -2.244 | 0.006 | 0.76 | -0.362 |
| 16 | -1.084 | -0.929 | 0.246 | -1.365 | 0.256 | 0.033 | 0.113 | 0.207 | 0.23 | 0.662 | 0.489 | 0.051 | -0.938 | -0.797 | 0.573 | -0.574 |
| 17 | -1.114 | 0.084 | 0.348 | -0.805 | -0.797 | 0.28 | -0.069 | 0.104 | 0.148 | 0.666 | 0.472 | 0.089 | -1.087 | -0.146 | 0.231 | -0.447 |
| 18 | -0.487 | -0.78 | -0.343 | -0.705 | 0.414 | 0.554 | -0.005 | -0.027 | 0.142 | 0.613 | 0.265 | -0.095 | -0.547 | 0.141 | 0.025 | -0.629 |
| 19 | 0.032 | -0.153 | 0.431 | -0.845 | 0.37 | 0.612 | -1.032 | 0.438 | -0.11 | 0.399 | 0.034 | -0.49 | -0.748 | 0.066 | -0.317 | -0.447 |
| 20 | -0.625 | 0.178 | 0.429 | -0.538 | 0.042 | 0.603 | 0.013 | 0.32 | -0.697 | 0.326 | 0.107 | -0.412 | -0.94 | -0.137 | 0.122 | -0.175 |
| 21 | -0.923 | -0.185 | -0.105 | -1.084 | -0.352 | 0.921 | 0.048 | 0.017 | -0.6 | 0.863 | -0.018 | -0.117 | -1.029 | 0.128 | 0.276 | -0.429 |
| 22 | -0.693 | -0.574 | -0.407 | -1.279 | 0.66 | 0.751 | -0.126 | 0.649 | -0.386 | 0.689 | 0.051 | -0.644 | -1.777 | -0.069 | 0.148 | -0.457 |