Model info
| Transcription factor | GATA1 | ||||||||
| Model | GATA1_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO |  | ||||||||
| LOGO (reverse complement) | 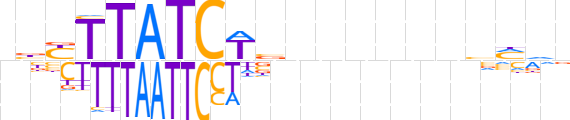 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 20 | ||||||||
| Quality | A | ||||||||
| Consensus | nbKbnnnnnnvWGATAARvn | ||||||||
| wAUC | 0.9053673015192157 | ||||||||
| Best AUC | 0.9489335987943514 | ||||||||
| Benchmark datasets | 12 | ||||||||
| Aligned words | 502 | ||||||||
| TF family | GATA-type zinc fingers{2.2.1} | ||||||||
| TF subfamily | Two zinc-finger GATA factors{2.2.1.1} | ||||||||
| HGNC | 4170 | ||||||||
| EntrezGene | 2623 | ||||||||
| UniProt ID | GATA1_HUMAN | ||||||||
| UniProt AC | P15976 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 19.708 | 19.683 | 17.377 | 28.657 | 27.596 | 19.766 | 1.277 | 119.989 | 17.519 | 36.566 | 14.924 | 56.627 | 4.3 | 27.101 | 67.81 | 21.1 |
| 02 | 9.909 | 4.773 | 36.155 | 18.285 | 35.718 | 5.362 | 8.062 | 53.975 | 23.397 | 13.267 | 37.82 | 26.904 | 0.905 | 3.928 | 199.251 | 22.289 |
| 03 | 15.715 | 12.961 | 39.483 | 1.769 | 5.969 | 5.621 | 4.033 | 11.707 | 44.834 | 81.198 | 83.282 | 71.975 | 7.926 | 10.811 | 91.908 | 10.807 |
| 04 | 24.822 | 4.786 | 33.797 | 11.038 | 51.501 | 16.909 | 4.412 | 37.769 | 40.881 | 37.541 | 79.998 | 60.287 | 13.913 | 12.41 | 49.552 | 20.383 |
| 05 | 37.107 | 22.989 | 54.184 | 16.837 | 24.231 | 22.102 | 5.88 | 19.432 | 30.295 | 51.781 | 47.397 | 38.287 | 19.174 | 29.642 | 52.088 | 28.574 |
| 06 | 29.636 | 15.502 | 55.869 | 9.8 | 37.978 | 36.964 | 7.051 | 44.521 | 32.953 | 39.565 | 58.882 | 28.149 | 13.438 | 22.494 | 49.906 | 17.292 |
| 07 | 31.05 | 18.937 | 52.764 | 11.253 | 37.275 | 25.563 | 11.546 | 40.14 | 36.427 | 39.015 | 63.928 | 32.339 | 15.573 | 18.88 | 39.155 | 26.154 |
| 08 | 36.0 | 22.576 | 47.002 | 14.748 | 43.388 | 23.763 | 3.901 | 31.343 | 44.286 | 47.33 | 47.558 | 28.219 | 11.41 | 26.677 | 45.333 | 26.467 |
| 09 | 36.64 | 12.141 | 60.731 | 25.571 | 57.509 | 32.08 | 6.799 | 23.958 | 50.69 | 27.513 | 44.243 | 21.348 | 21.805 | 20.262 | 37.849 | 20.861 |
| 10 | 28.841 | 66.917 | 62.486 | 8.4 | 21.191 | 48.295 | 2.952 | 19.557 | 18.386 | 64.862 | 48.184 | 18.189 | 4.927 | 37.225 | 39.488 | 10.099 |
| 11 | 57.276 | 3.898 | 1.038 | 11.132 | 122.378 | 3.023 | 0.895 | 91.004 | 117.888 | 2.031 | 0.0 | 33.191 | 38.301 | 0.888 | 0.901 | 16.155 |
| 12 | 0.0 | 0.0 | 335.843 | 0.0 | 0.0 | 0.0 | 9.841 | 0.0 | 0.0 | 0.0 | 2.834 | 0.0 | 0.0 | 0.0 | 151.483 | 0.0 |
| 13 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 500.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 14 | 0.0 | 4.787 | 3.224 | 491.989 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 15 | 0.0 | 0.0 | 0.0 | 0.0 | 4.787 | 0.0 | 0.0 | 0.0 | 2.135 | 1.089 | 0.0 | 0.0 | 485.404 | 0.0 | 0.0 | 6.584 |
| 16 | 445.469 | 0.0 | 31.058 | 15.8 | 1.089 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 6.584 | 0.0 | 0.0 | 0.0 |
| 17 | 74.485 | 64.817 | 293.556 | 20.284 | 0.0 | 0.0 | 0.0 | 0.0 | 4.065 | 8.106 | 16.814 | 2.073 | 0.0 | 0.0 | 15.8 | 0.0 |
| 18 | 23.62 | 12.965 | 39.993 | 1.972 | 41.374 | 13.557 | 9.838 | 8.154 | 106.893 | 90.763 | 115.738 | 12.776 | 2.837 | 2.821 | 13.661 | 3.038 |
| 19 | 54.602 | 25.424 | 64.364 | 30.334 | 59.698 | 22.516 | 7.223 | 30.668 | 60.261 | 43.226 | 42.033 | 33.71 | 6.892 | 4.724 | 11.414 | 2.911 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.454 | -0.455 | -0.577 | -0.086 | -0.123 | -0.451 | -2.944 | 1.336 | -0.569 | 0.155 | -0.726 | 0.589 | -1.909 | -0.141 | 0.768 | -0.387 |
| 02 | -1.122 | -1.813 | 0.144 | -0.527 | 0.132 | -1.705 | -1.32 | 0.541 | -0.285 | -0.84 | 0.189 | -0.148 | -3.197 | -1.992 | 1.842 | -0.333 |
| 03 | -0.675 | -0.863 | 0.231 | -2.685 | -1.605 | -1.661 | -1.968 | -0.962 | 0.357 | 0.947 | 0.973 | 0.827 | -1.336 | -1.038 | 1.071 | -1.039 |
| 04 | -0.227 | -1.811 | 0.077 | -1.018 | 0.495 | -0.604 | -1.886 | 0.187 | 0.266 | 0.181 | 0.932 | 0.651 | -0.794 | -0.905 | 0.456 | -0.421 |
| 05 | 0.17 | -0.303 | 0.545 | -0.608 | -0.251 | -0.341 | -1.619 | -0.468 | -0.031 | 0.5 | 0.412 | 0.201 | -0.481 | -0.052 | 0.506 | -0.088 |
| 06 | -0.052 | -0.689 | 0.576 | -1.133 | 0.193 | 0.166 | -1.448 | 0.35 | 0.052 | 0.233 | 0.628 | -0.103 | -0.828 | -0.324 | 0.464 | -0.582 |
| 07 | -0.006 | -0.493 | 0.519 | -1.0 | 0.174 | -0.198 | -0.975 | 0.248 | 0.152 | 0.219 | 0.709 | 0.034 | -0.684 | -0.496 | 0.223 | -0.176 |
| 08 | 0.14 | -0.32 | 0.404 | -0.737 | 0.325 | -0.27 | -1.998 | 0.003 | 0.345 | 0.411 | 0.416 | -0.101 | -0.986 | -0.156 | 0.368 | -0.164 |
| 09 | 0.157 | -0.926 | 0.658 | -0.198 | 0.604 | 0.026 | -1.482 | -0.262 | 0.479 | -0.126 | 0.344 | -0.375 | -0.355 | -0.427 | 0.189 | -0.398 |
| 10 | -0.079 | 0.755 | 0.687 | -1.281 | -0.383 | 0.431 | -2.248 | -0.461 | -0.522 | 0.724 | 0.429 | -0.532 | -1.784 | 0.173 | 0.231 | -1.104 |
| 11 | 0.6 | -1.999 | -3.099 | -1.01 | 1.356 | -2.227 | -3.205 | 1.061 | 1.319 | -2.571 | -4.4 | 0.06 | 0.201 | -3.21 | -3.2 | -0.648 |
| 12 | -4.4 | -4.4 | 2.363 | -4.4 | -4.4 | -4.4 | -1.129 | -4.4 | -4.4 | -4.4 | -2.284 | -4.4 | -4.4 | -4.4 | 1.569 | -4.4 |
| 13 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | 2.761 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 |
| 14 | -4.4 | -1.81 | -2.17 | 2.745 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 |
| 15 | -4.4 | -4.4 | -4.4 | -4.4 | -1.81 | -4.4 | -4.4 | -4.4 | -2.529 | -3.064 | -4.4 | -4.4 | 2.731 | -4.4 | -4.4 | -1.512 |
| 16 | 2.646 | -4.4 | -0.006 | -0.67 | -3.064 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -1.512 | -4.4 | -4.4 | -4.4 |
| 17 | 0.861 | 0.723 | 2.229 | -0.426 | -4.4 | -4.4 | -4.4 | -4.4 | -1.961 | -1.315 | -0.609 | -2.554 | -4.4 | -4.4 | -0.67 | -4.4 |
| 18 | -0.276 | -0.863 | 0.244 | -2.596 | 0.278 | -0.819 | -1.129 | -1.309 | 1.221 | 1.058 | 1.3 | -0.877 | -2.283 | -2.288 | -0.812 | -2.223 |
| 19 | 0.553 | -0.204 | 0.716 | -0.029 | 0.641 | -0.323 | -1.425 | -0.019 | 0.651 | 0.321 | 0.293 | 0.075 | -1.469 | -1.823 | -0.986 | -2.261 |