Model info
| Transcription factor | Fli1 | ||||||||
| Model | FLI1_MOUSE.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 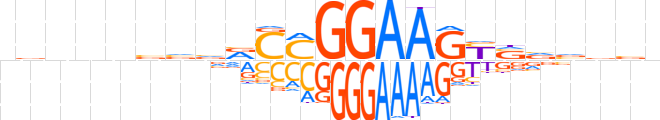 | ||||||||
| LOGO (reverse complement) | 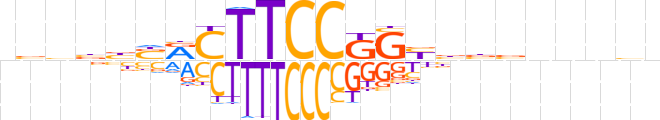 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Consensus | nbnnnvvvRCMGGAAGYSvvdbn | ||||||||
| wAUC | 0.8591646810780967 | ||||||||
| Best AUC | 0.9329776164245106 | ||||||||
| Benchmark datasets | 3 | ||||||||
| Aligned words | 457 | ||||||||
| TF family | Ets-related factors{3.5.2} | ||||||||
| TF subfamily | Ets-like factors{3.5.2.1} | ||||||||
| MGI | 95554 | ||||||||
| EntrezGene | 14247 | ||||||||
| UniProt ID | FLI1_MOUSE | ||||||||
| UniProt AC | P26323 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 10.03 | 18.868 | 41.941 | 10.689 | 23.442 | 39.468 | 34.717 | 26.5 | 25.704 | 25.733 | 58.344 | 14.632 | 2.999 | 17.791 | 26.988 | 19.417 |
| 02 | 17.127 | 16.335 | 21.746 | 6.967 | 17.837 | 21.048 | 31.032 | 31.944 | 30.661 | 41.323 | 65.512 | 24.495 | 14.162 | 21.562 | 21.602 | 13.912 |
| 03 | 21.779 | 17.809 | 38.19 | 2.008 | 19.051 | 37.987 | 17.023 | 26.207 | 37.989 | 49.015 | 35.683 | 17.205 | 11.246 | 23.987 | 18.983 | 23.102 |
| 04 | 33.426 | 9.495 | 35.991 | 11.153 | 20.42 | 43.535 | 40.483 | 24.36 | 26.166 | 44.563 | 28.002 | 11.149 | 12.31 | 21.372 | 24.742 | 10.1 |
| 05 | 16.036 | 25.454 | 28.264 | 22.568 | 18.652 | 40.701 | 46.767 | 12.845 | 15.605 | 46.435 | 58.556 | 8.621 | 2.095 | 27.494 | 19.158 | 8.013 |
| 06 | 11.609 | 22.993 | 15.968 | 1.817 | 41.373 | 47.367 | 38.535 | 12.81 | 34.561 | 44.257 | 61.211 | 12.716 | 8.997 | 11.201 | 30.957 | 0.892 |
| 07 | 28.658 | 19.874 | 45.074 | 2.934 | 43.337 | 31.058 | 31.497 | 19.927 | 44.971 | 34.562 | 59.439 | 7.699 | 1.81 | 13.74 | 11.758 | 0.926 |
| 08 | 66.528 | 9.913 | 37.327 | 5.008 | 44.092 | 16.022 | 31.548 | 7.572 | 94.945 | 7.808 | 41.89 | 3.126 | 8.708 | 4.7 | 9.009 | 9.068 |
| 09 | 20.282 | 169.128 | 24.863 | 0.0 | 0.0 | 37.383 | 1.06 | 0.0 | 2.906 | 93.195 | 22.656 | 1.016 | 0.833 | 22.102 | 1.838 | 0.0 |
| 10 | 4.942 | 7.098 | 0.916 | 11.065 | 77.537 | 239.3 | 3.071 | 1.901 | 21.402 | 27.997 | 0.0 | 1.019 | 0.0 | 1.016 | 0.0 | 0.0 |
| 11 | 0.885 | 0.861 | 101.248 | 0.887 | 0.0 | 0.0 | 275.412 | 0.0 | 0.0 | 0.0 | 3.987 | 0.0 | 0.0 | 0.0 | 13.984 | 0.0 |
| 12 | 0.0 | 0.0 | 0.885 | 0.0 | 0.861 | 0.0 | 0.0 | 0.0 | 0.912 | 0.0 | 393.719 | 0.0 | 0.0 | 0.0 | 0.887 | 0.0 |
| 13 | 1.773 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 394.554 | 0.937 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 14 | 380.64 | 0.0 | 1.832 | 13.855 | 0.937 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 15 | 57.017 | 5.69 | 308.757 | 10.112 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.832 | 0.0 | 0.96 | 0.0 | 12.896 | 0.0 |
| 16 | 4.774 | 20.667 | 3.96 | 28.576 | 0.0 | 2.869 | 0.0 | 2.821 | 18.583 | 68.616 | 13.869 | 222.417 | 0.0 | 0.871 | 8.409 | 0.833 |
| 17 | 2.868 | 3.813 | 16.677 | 0.0 | 23.075 | 13.902 | 35.019 | 21.026 | 3.082 | 2.075 | 20.115 | 0.966 | 20.842 | 35.373 | 174.343 | 24.089 |
| 18 | 7.301 | 2.821 | 37.635 | 2.11 | 7.833 | 21.082 | 14.711 | 11.536 | 78.896 | 33.326 | 120.607 | 13.326 | 4.123 | 7.944 | 29.066 | 4.946 |
| 19 | 21.602 | 47.814 | 26.813 | 1.925 | 10.779 | 31.338 | 13.538 | 9.517 | 47.946 | 76.28 | 57.95 | 19.843 | 1.921 | 5.058 | 21.901 | 3.037 |
| 20 | 28.267 | 11.931 | 25.107 | 16.943 | 25.857 | 21.933 | 79.948 | 32.754 | 20.866 | 25.744 | 49.425 | 24.168 | 3.063 | 7.956 | 9.863 | 13.441 |
| 21 | 14.498 | 10.351 | 44.577 | 8.627 | 9.587 | 16.714 | 26.6 | 14.663 | 26.042 | 56.957 | 58.021 | 23.322 | 7.123 | 13.769 | 49.726 | 16.687 |
| 22 | 12.187 | 18.519 | 16.467 | 10.077 | 24.141 | 32.09 | 23.575 | 17.984 | 32.693 | 51.331 | 60.27 | 34.629 | 5.117 | 18.918 | 22.553 | 16.712 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -0.885 | -0.27 | 0.518 | -0.823 | -0.057 | 0.458 | 0.331 | 0.064 | 0.034 | 0.035 | 0.846 | -0.518 | -2.011 | -0.327 | 0.082 | -0.242 |
| 02 | -0.365 | -0.411 | -0.13 | -1.233 | -0.325 | -0.163 | 0.22 | 0.249 | 0.208 | 0.503 | 0.961 | -0.013 | -0.55 | -0.139 | -0.137 | -0.568 |
| 03 | -0.129 | -0.326 | 0.425 | -2.359 | -0.26 | 0.42 | -0.371 | 0.053 | 0.42 | 0.673 | 0.358 | -0.36 | -0.774 | -0.034 | -0.264 | -0.071 |
| 04 | 0.293 | -0.938 | 0.367 | -0.782 | -0.192 | 0.555 | 0.483 | -0.019 | 0.052 | 0.578 | 0.119 | -0.783 | -0.687 | -0.148 | -0.003 | -0.878 |
| 05 | -0.429 | 0.025 | 0.128 | -0.094 | -0.281 | 0.488 | 0.626 | -0.645 | -0.456 | 0.619 | 0.849 | -1.03 | -2.323 | 0.101 | -0.255 | -1.1 |
| 06 | -0.743 | -0.076 | -0.433 | -2.443 | 0.505 | 0.639 | 0.434 | -0.648 | 0.327 | 0.571 | 0.893 | -0.655 | -0.989 | -0.778 | 0.218 | -2.991 |
| 07 | 0.141 | -0.219 | 0.59 | -2.031 | 0.551 | 0.221 | 0.235 | -0.216 | 0.587 | 0.327 | 0.864 | -1.138 | -2.446 | -0.58 | -0.731 | -2.965 |
| 08 | 0.976 | -0.896 | 0.403 | -1.544 | 0.568 | -0.43 | 0.236 | -1.154 | 1.33 | -1.125 | 0.517 | -1.974 | -1.021 | -1.603 | -0.988 | -0.982 |
| 09 | -0.199 | 1.906 | 0.001 | -4.21 | -4.21 | 0.404 | -2.866 | -4.21 | -2.039 | 1.312 | -0.09 | -2.897 | -3.039 | -0.115 | -2.433 | -4.21 |
| 10 | -1.556 | -1.216 | -2.972 | -0.79 | 1.129 | 2.252 | -1.99 | -2.405 | -0.146 | 0.118 | -4.21 | -2.896 | -4.21 | -2.897 | -4.21 | -4.21 |
| 11 | -2.997 | -3.016 | 1.394 | -2.995 | -4.21 | -4.21 | 2.393 | -4.21 | -4.21 | -4.21 | -1.754 | -4.21 | -4.21 | -4.21 | -0.563 | -4.21 |
| 12 | -4.21 | -4.21 | -2.997 | -4.21 | -3.016 | -4.21 | -4.21 | -4.21 | -2.975 | -4.21 | 2.75 | -4.21 | -4.21 | -4.21 | -2.995 | -4.21 |
| 13 | -2.463 | -4.21 | -4.21 | -4.21 | -4.21 | -4.21 | -4.21 | -4.21 | 2.752 | -2.956 | -4.21 | -4.21 | -4.21 | -4.21 | -4.21 | -4.21 |
| 14 | 2.716 | -4.21 | -2.436 | -0.572 | -2.956 | -4.21 | -4.21 | -4.21 | -4.21 | -4.21 | -4.21 | -4.21 | -4.21 | -4.21 | -4.21 | -4.21 |
| 15 | 0.823 | -1.425 | 2.507 | -0.877 | -4.21 | -4.21 | -4.21 | -4.21 | -4.21 | -4.21 | -2.436 | -4.21 | -2.939 | -4.21 | -0.641 | -4.21 |
| 16 | -1.588 | -0.18 | -1.761 | 0.139 | -4.21 | -2.05 | -4.21 | -2.065 | -0.285 | 1.007 | -0.571 | 2.179 | -4.21 | -3.008 | -1.054 | -3.039 |
| 17 | -2.051 | -1.795 | -0.391 | -4.21 | -0.072 | -0.568 | 0.34 | -0.164 | -1.987 | -2.331 | -0.207 | -2.934 | -0.172 | 0.349 | 1.936 | -0.03 |
| 18 | -1.189 | -2.066 | 0.411 | -2.317 | -1.122 | -0.161 | -0.513 | -0.75 | 1.146 | 0.291 | 1.569 | -0.61 | -1.723 | -1.109 | 0.155 | -1.555 |
| 19 | -0.137 | 0.648 | 0.076 | -2.394 | -0.815 | 0.23 | -0.594 | -0.935 | 0.651 | 1.112 | 0.839 | -0.22 | -2.396 | -1.535 | -0.123 | -2.0 |
| 20 | 0.128 | -0.717 | 0.011 | -0.375 | 0.04 | -0.122 | 1.159 | 0.273 | -0.171 | 0.036 | 0.681 | -0.027 | -1.992 | -1.107 | -0.901 | -0.601 |
| 21 | -0.527 | -0.854 | 0.579 | -1.03 | -0.928 | -0.389 | 0.068 | -0.516 | 0.047 | 0.822 | 0.84 | -0.062 | -1.212 | -0.578 | 0.687 | -0.39 |
| 22 | -0.696 | -0.288 | -0.403 | -0.88 | -0.028 | 0.253 | -0.051 | -0.317 | 0.272 | 0.719 | 0.878 | 0.328 | -1.524 | -0.267 | -0.095 | -0.389 |