Model info
| Transcription factor | ETV1 | ||||||||
| Model | ETV1_HUMAN.H10MO.B | ||||||||
| Model type | Mononucleotide PWM | ||||||||
| LOGO | 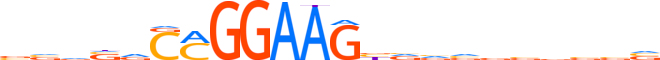 | ||||||||
| LOGO (reverse complement) |  | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 22 | ||||||||
| Quality | B | ||||||||
| Consensus | vvvvvCMGGAAGbvvvvvbbvv | ||||||||
| wAUC | 0.756638256277947 | ||||||||
| Best AUC | 0.8013240919289543 | ||||||||
| Benchmark datasets | 2 | ||||||||
| Aligned words | 339 | ||||||||
| TF family | Ets-related factors{3.5.2} | ||||||||
| TF subfamily | Elk-like factors{3.5.2.2} | ||||||||
| HGNC | 3490 | ||||||||
| EntrezGene | 2115 | ||||||||
| UniProt ID | ETV1_HUMAN | ||||||||
| UniProt AC | P50549 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | 57.539 | 128.994 | 116.715 | 33.793 |
| 02 | 69.256 | 85.305 | 156.681 | 25.798 |
| 03 | 95.224 | 82.451 | 125.62 | 33.744 |
| 04 | 42.972 | 72.381 | 186.008 | 35.678 |
| 05 | 119.388 | 40.504 | 143.227 | 33.922 |
| 06 | 20.077 | 274.934 | 42.029 | 0.0 |
| 07 | 142.597 | 192.457 | 1.985 | 0.0 |
| 08 | 0.0 | 0.884 | 336.155 | 0.0 |
| 09 | 0.0 | 0.0 | 337.04 | 0.0 |
| 10 | 334.948 | 1.003 | 1.089 | 0.0 |
| 11 | 326.826 | 0.0 | 0.0 | 10.214 |
| 12 | 43.315 | 1.98 | 289.126 | 2.619 |
| 13 | 26.192 | 127.59 | 53.168 | 130.089 |
| 14 | 90.663 | 52.995 | 167.551 | 25.831 |
| 15 | 100.157 | 70.959 | 140.101 | 25.822 |
| 16 | 89.169 | 112.285 | 115.829 | 19.756 |
| 17 | 65.765 | 78.252 | 143.581 | 49.441 |
| 18 | 48.871 | 103.5 | 137.405 | 47.263 |
| 19 | 47.186 | 96.961 | 126.683 | 66.21 |
| 20 | 46.042 | 84.677 | 147.464 | 58.857 |
| 21 | 51.554 | 85.625 | 153.569 | 46.291 |
| 22 | 64.306 | 116.368 | 140.207 | 16.159 |
PWM
| A | C | G | T | |
|---|---|---|---|---|
| 01 | -0.374 | 0.42 | 0.321 | -0.889 |
| 02 | -0.192 | 0.012 | 0.612 | -1.146 |
| 03 | 0.12 | -0.021 | 0.394 | -0.89 |
| 04 | -0.657 | -0.149 | 0.783 | -0.837 |
| 05 | 0.343 | -0.714 | 0.524 | -0.885 |
| 06 | -1.381 | 1.171 | -0.679 | -4.076 |
| 07 | 0.519 | 0.816 | -3.215 | -4.076 |
| 08 | -4.076 | -3.601 | 1.371 | -4.076 |
| 09 | -4.076 | -4.076 | 1.373 | -4.076 |
| 10 | 1.367 | -3.552 | -3.517 | -4.076 |
| 11 | 1.343 | -4.076 | -4.076 | -1.994 |
| 12 | -0.649 | -3.217 | 1.221 | -3.046 |
| 13 | -1.131 | 0.409 | -0.451 | 0.428 |
| 14 | 0.072 | -0.454 | 0.679 | -1.145 |
| 15 | 0.17 | -0.169 | 0.502 | -1.145 |
| 16 | 0.056 | 0.283 | 0.314 | -1.396 |
| 17 | -0.243 | -0.073 | 0.526 | -0.521 |
| 18 | -0.532 | 0.203 | 0.482 | -0.565 |
| 19 | -0.567 | 0.138 | 0.402 | -0.236 |
| 20 | -0.59 | 0.005 | 0.552 | -0.351 |
| 21 | -0.481 | 0.016 | 0.593 | -0.585 |
| 22 | -0.265 | 0.318 | 0.502 | -1.582 |