Model info
| Transcription factor | ETV1 | ||||||||
| Model | ETV1_HUMAN.H10DI.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 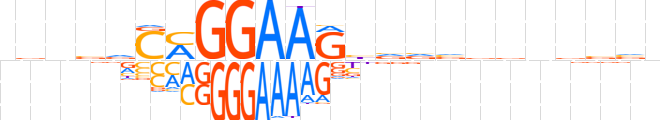 | ||||||||
| LOGO (reverse complement) | 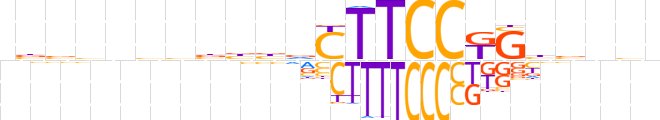 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | B | ||||||||
| Consensus | nvnvvCMGGAAGbvvvvvnbbvn | ||||||||
| wAUC | 0.7847037935290183 | ||||||||
| Best AUC | 0.843438278254048 | ||||||||
| Benchmark datasets | 2 | ||||||||
| Aligned words | 433 | ||||||||
| TF family | Ets-related factors{3.5.2} | ||||||||
| TF subfamily | Elk-like factors{3.5.2.2} | ||||||||
| HGNC | 3490 | ||||||||
| EntrezGene | 2115 | ||||||||
| UniProt ID | ETV1_HUMAN | ||||||||
| UniProt AC | P50549 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 32.989 | 7.002 | 36.822 | 11.679 | 44.542 | 34.489 | 42.682 | 27.594 | 31.223 | 31.718 | 62.96 | 7.912 | 2.719 | 27.394 | 23.057 | 7.252 |
| 02 | 39.872 | 7.757 | 50.412 | 13.431 | 25.771 | 30.957 | 18.014 | 25.862 | 53.114 | 46.833 | 53.038 | 12.535 | 6.244 | 19.333 | 22.658 | 6.201 |
| 03 | 30.684 | 22.638 | 60.59 | 11.089 | 9.103 | 22.839 | 42.64 | 30.298 | 29.028 | 42.211 | 65.965 | 6.918 | 3.317 | 11.093 | 28.699 | 14.921 |
| 04 | 30.731 | 4.738 | 31.122 | 5.541 | 46.511 | 14.79 | 19.92 | 17.561 | 64.843 | 26.987 | 89.291 | 16.773 | 12.269 | 20.019 | 18.455 | 12.484 |
| 05 | 12.781 | 114.57 | 27.001 | 0.0 | 2.766 | 63.768 | 0.0 | 0.0 | 9.661 | 126.205 | 21.756 | 1.166 | 0.881 | 49.4 | 2.077 | 0.0 |
| 06 | 20.124 | 4.006 | 1.02 | 0.939 | 171.491 | 179.561 | 1.085 | 1.806 | 28.276 | 22.558 | 0.0 | 0.0 | 1.166 | 0.0 | 0.0 | 0.0 |
| 07 | 0.0 | 1.036 | 220.022 | 0.0 | 0.0 | 0.0 | 206.125 | 0.0 | 0.0 | 0.0 | 2.105 | 0.0 | 0.0 | 0.0 | 2.745 | 0.0 |
| 08 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.036 | 0.0 | 0.0 | 0.0 | 430.997 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 09 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 431.03 | 1.003 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 10 | 414.753 | 0.0 | 1.134 | 15.143 | 1.003 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 11 | 69.311 | 14.449 | 330.804 | 1.192 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.134 | 0.0 | 0.984 | 0.0 | 14.158 | 0.0 |
| 12 | 5.983 | 24.665 | 8.297 | 31.351 | 5.271 | 4.638 | 0.0 | 4.54 | 29.913 | 116.787 | 68.704 | 130.693 | 0.0 | 0.0 | 0.0 | 1.192 |
| 13 | 7.023 | 5.754 | 26.357 | 2.033 | 53.108 | 23.531 | 54.552 | 14.899 | 41.165 | 10.679 | 21.79 | 3.366 | 23.601 | 25.428 | 97.263 | 21.485 |
| 14 | 46.358 | 13.824 | 56.464 | 8.251 | 26.105 | 18.869 | 13.848 | 6.57 | 50.169 | 48.723 | 89.799 | 11.272 | 5.417 | 7.922 | 21.652 | 6.791 |
| 15 | 31.199 | 29.932 | 59.842 | 7.075 | 27.262 | 28.775 | 18.445 | 14.855 | 56.134 | 66.594 | 50.32 | 8.715 | 7.962 | 11.739 | 13.184 | 0.0 |
| 16 | 31.265 | 17.108 | 61.009 | 13.176 | 18.031 | 33.977 | 52.626 | 32.405 | 27.37 | 43.205 | 46.271 | 24.945 | 4.734 | 7.66 | 13.497 | 4.753 |
| 17 | 23.407 | 11.035 | 39.108 | 7.85 | 12.845 | 36.463 | 20.474 | 32.168 | 32.042 | 59.892 | 64.723 | 16.746 | 1.915 | 17.462 | 44.664 | 11.238 |
| 18 | 15.827 | 9.654 | 39.076 | 5.652 | 16.728 | 44.613 | 29.989 | 33.523 | 41.182 | 58.566 | 40.97 | 28.251 | 8.523 | 21.844 | 26.354 | 11.28 |
| 19 | 16.928 | 12.28 | 43.802 | 9.251 | 22.623 | 44.838 | 35.907 | 31.31 | 24.016 | 36.637 | 57.306 | 18.429 | 2.057 | 14.677 | 43.057 | 18.916 |
| 20 | 10.097 | 9.713 | 33.617 | 12.197 | 23.377 | 28.083 | 33.132 | 23.839 | 18.982 | 39.494 | 103.79 | 17.806 | 10.023 | 22.046 | 26.974 | 18.863 |
| 21 | 17.821 | 11.869 | 23.582 | 9.207 | 29.198 | 39.416 | 22.777 | 7.944 | 35.73 | 64.8 | 85.888 | 11.095 | 11.431 | 28.713 | 28.032 | 4.529 |
| 22 | 34.305 | 13.019 | 36.599 | 10.257 | 26.536 | 43.694 | 36.71 | 37.858 | 38.444 | 40.918 | 55.987 | 24.931 | 1.007 | 9.949 | 16.839 | 4.981 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.198 | -1.311 | 0.306 | -0.82 | 0.495 | 0.242 | 0.453 | 0.021 | 0.143 | 0.159 | 0.839 | -1.195 | -2.179 | 0.014 | -0.156 | -1.278 |
| 02 | 0.385 | -1.213 | 0.618 | -0.684 | -0.046 | 0.135 | -0.398 | -0.043 | 0.67 | 0.545 | 0.668 | -0.752 | -1.419 | -0.329 | -0.173 | -1.426 |
| 03 | 0.126 | -0.174 | 0.801 | -0.87 | -1.06 | -0.165 | 0.452 | 0.114 | 0.071 | 0.442 | 0.885 | -1.322 | -2.003 | -0.87 | 0.06 | -0.582 |
| 04 | 0.128 | -1.677 | 0.14 | -1.532 | 0.538 | -0.591 | -0.299 | -0.423 | 0.868 | -0.001 | 1.186 | -0.468 | -0.772 | -0.294 | -0.374 | -0.756 |
| 05 | -0.733 | 1.435 | -0.0 | -4.279 | -2.164 | 0.851 | -4.279 | -4.279 | -1.003 | 1.531 | -0.213 | -2.875 | -3.079 | 0.598 | -2.411 | -4.279 |
| 06 | -0.289 | -1.832 | -2.974 | -3.034 | 1.837 | 1.883 | -2.929 | -2.528 | 0.045 | -0.177 | -4.279 | -4.279 | -2.875 | -4.279 | -4.279 | -4.279 |
| 07 | -4.279 | -2.962 | 2.086 | -4.279 | -4.279 | -4.279 | 2.02 | -4.279 | -4.279 | -4.279 | -2.4 | -4.279 | -4.279 | -4.279 | -2.171 | -4.279 |
| 08 | -4.279 | -4.279 | -4.279 | -4.279 | -4.279 | -4.279 | -2.962 | -4.279 | -4.279 | -4.279 | 2.757 | -4.279 | -4.279 | -4.279 | -4.279 | -4.279 |
| 09 | -4.279 | -4.279 | -4.279 | -4.279 | -4.279 | -4.279 | -4.279 | -4.279 | 2.757 | -2.986 | -4.279 | -4.279 | -4.279 | -4.279 | -4.279 | -4.279 |
| 10 | 2.719 | -4.279 | -2.896 | -0.568 | -2.986 | -4.279 | -4.279 | -4.279 | -4.279 | -4.279 | -4.279 | -4.279 | -4.279 | -4.279 | -4.279 | -4.279 |
| 11 | 0.934 | -0.613 | 2.493 | -2.858 | -4.279 | -4.279 | -4.279 | -4.279 | -4.279 | -4.279 | -2.896 | -4.279 | -3.0 | -4.279 | -0.633 | -4.279 |
| 12 | -1.459 | -0.089 | -1.149 | 0.147 | -1.578 | -1.697 | -4.279 | -1.717 | 0.101 | 1.454 | 0.925 | 1.566 | -4.279 | -4.279 | -4.279 | -2.858 |
| 13 | -1.308 | -1.496 | -0.024 | -2.429 | 0.67 | -0.136 | 0.696 | -0.583 | 0.417 | -0.907 | -0.211 | -1.989 | -0.133 | -0.059 | 1.271 | -0.225 |
| 14 | 0.535 | -0.656 | 0.73 | -1.155 | -0.033 | -0.352 | -0.655 | -1.371 | 0.613 | 0.584 | 1.192 | -0.854 | -1.553 | -1.194 | -0.217 | -1.34 |
| 15 | 0.143 | 0.102 | 0.788 | -1.301 | 0.009 | 0.063 | -0.375 | -0.586 | 0.725 | 0.894 | 0.616 | -1.102 | -1.189 | -0.815 | -0.703 | -4.279 |
| 16 | 0.145 | -0.448 | 0.807 | -0.703 | -0.397 | 0.227 | 0.661 | 0.18 | 0.013 | 0.465 | 0.533 | -0.078 | -1.678 | -1.225 | -0.68 | -1.674 |
| 17 | -0.141 | -0.875 | 0.366 | -1.202 | -0.728 | 0.297 | -0.272 | 0.173 | 0.169 | 0.789 | 0.866 | -0.469 | -2.479 | -0.428 | 0.498 | -0.857 |
| 18 | -0.524 | -1.004 | 0.365 | -1.513 | -0.47 | 0.497 | 0.104 | 0.214 | 0.417 | 0.767 | 0.412 | 0.045 | -1.124 | -0.209 | -0.024 | -0.854 |
| 19 | -0.459 | -0.771 | 0.478 | -1.045 | -0.174 | 0.502 | 0.282 | 0.146 | -0.115 | 0.301 | 0.745 | -0.376 | -2.42 | -0.598 | 0.461 | -0.35 |
| 20 | -0.961 | -0.998 | 0.216 | -0.778 | -0.142 | 0.039 | 0.202 | -0.123 | -0.347 | 0.376 | 1.336 | -0.409 | -0.968 | -0.2 | -0.001 | -0.353 |
| 21 | -0.408 | -0.804 | -0.133 | -1.05 | 0.077 | 0.374 | -0.168 | -1.191 | 0.277 | 0.867 | 1.148 | -0.87 | -0.841 | 0.061 | 0.037 | -1.719 |
| 22 | 0.236 | -0.715 | 0.3 | -0.946 | -0.017 | 0.476 | 0.303 | 0.334 | 0.349 | 0.411 | 0.722 | -0.079 | -2.983 | -0.975 | -0.464 | -1.631 |