Model info
| Transcription factor | ERG | ||||||||
| Model | ERG_HUMAN.H10DI.B | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 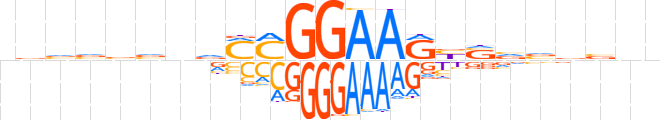 | ||||||||
| LOGO (reverse complement) | 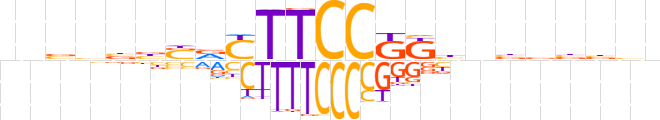 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | B | ||||||||
| Consensus | vvvvbvvvMMGGAARbSvvbbnn | ||||||||
| wAUC | 0.8405842474173834 | ||||||||
| Best AUC | 0.8950081717012444 | ||||||||
| Benchmark datasets | 2 | ||||||||
| Aligned words | 497 | ||||||||
| TF family | Ets-related factors{3.5.2} | ||||||||
| TF subfamily | Ets-like factors{3.5.2.1} | ||||||||
| HGNC | 3446 | ||||||||
| EntrezGene | 2078 | ||||||||
| UniProt ID | ERG_HUMAN | ||||||||
| UniProt AC | P11308 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 8.662 | 21.537 | 32.554 | 21.405 | 45.377 | 31.451 | 55.646 | 19.618 | 27.752 | 47.742 | 79.861 | 18.169 | 10.974 | 32.324 | 27.035 | 5.893 |
| 02 | 19.696 | 36.325 | 31.865 | 4.88 | 28.344 | 55.398 | 27.461 | 21.851 | 47.429 | 85.227 | 42.48 | 19.96 | 10.66 | 31.962 | 16.698 | 5.763 |
| 03 | 34.002 | 24.864 | 42.141 | 5.121 | 55.798 | 84.468 | 48.965 | 19.682 | 15.943 | 44.732 | 47.028 | 10.801 | 5.901 | 15.854 | 24.863 | 5.836 |
| 04 | 23.329 | 28.92 | 28.091 | 31.304 | 30.37 | 65.591 | 52.377 | 21.579 | 11.512 | 65.182 | 62.089 | 24.215 | 0.99 | 15.59 | 19.879 | 4.982 |
| 05 | 8.526 | 31.538 | 25.119 | 1.019 | 34.257 | 67.594 | 54.427 | 19.004 | 44.486 | 53.12 | 56.016 | 8.814 | 2.797 | 26.799 | 41.463 | 11.022 |
| 06 | 21.314 | 18.514 | 43.211 | 7.027 | 36.855 | 42.075 | 49.64 | 50.479 | 24.465 | 62.555 | 75.17 | 14.836 | 2.89 | 19.698 | 12.305 | 4.967 |
| 07 | 37.513 | 24.388 | 21.7 | 1.923 | 43.337 | 19.713 | 57.217 | 22.576 | 74.126 | 18.428 | 79.739 | 8.033 | 18.907 | 17.029 | 30.687 | 10.686 |
| 08 | 27.396 | 126.403 | 14.745 | 5.338 | 12.995 | 61.54 | 1.913 | 3.109 | 10.447 | 138.333 | 26.398 | 14.166 | 0.0 | 37.355 | 3.805 | 2.057 |
| 09 | 23.476 | 19.783 | 0.0 | 7.578 | 88.774 | 270.767 | 2.142 | 1.947 | 11.658 | 35.204 | 0.0 | 0.0 | 12.585 | 3.143 | 1.053 | 7.889 |
| 10 | 2.928 | 1.14 | 132.425 | 0.0 | 1.036 | 0.975 | 326.887 | 0.0 | 0.0 | 0.0 | 3.195 | 0.0 | 0.0 | 0.0 | 17.415 | 0.0 |
| 11 | 0.0 | 0.0 | 3.964 | 0.0 | 0.0 | 0.975 | 1.14 | 0.0 | 0.92 | 0.0 | 479.001 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 12 | 0.92 | 0.0 | 0.0 | 0.0 | 0.975 | 0.0 | 0.0 | 0.0 | 461.164 | 21.764 | 1.177 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 13 | 445.287 | 2.86 | 2.928 | 11.985 | 18.563 | 1.203 | 1.027 | 0.971 | 1.177 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 14 | 89.167 | 12.784 | 339.569 | 23.507 | 0.0 | 0.0 | 4.063 | 0.0 | 1.119 | 0.0 | 2.836 | 0.0 | 1.023 | 0.0 | 11.932 | 0.0 |
| 15 | 1.005 | 49.148 | 4.954 | 36.203 | 2.991 | 2.089 | 0.0 | 7.703 | 31.479 | 108.614 | 29.393 | 188.913 | 0.0 | 3.085 | 13.233 | 7.189 |
| 16 | 4.93 | 5.97 | 24.576 | 0.0 | 25.624 | 30.372 | 83.429 | 23.511 | 16.58 | 0.0 | 29.068 | 1.931 | 17.006 | 31.606 | 170.068 | 21.329 |
| 17 | 21.096 | 8.678 | 31.322 | 3.044 | 21.933 | 16.344 | 24.787 | 4.884 | 102.881 | 69.662 | 121.52 | 13.077 | 12.428 | 15.996 | 14.621 | 3.725 |
| 18 | 47.5 | 69.878 | 35.08 | 5.881 | 15.724 | 41.83 | 37.901 | 15.225 | 32.709 | 88.53 | 60.026 | 10.985 | 4.939 | 10.098 | 9.694 | 0.0 |
| 19 | 18.933 | 1.953 | 41.369 | 38.619 | 18.869 | 39.595 | 88.63 | 63.242 | 29.012 | 41.306 | 47.918 | 24.465 | 0.0 | 12.813 | 13.239 | 6.04 |
| 20 | 7.27 | 6.911 | 44.1 | 8.533 | 12.958 | 25.462 | 37.627 | 19.619 | 24.833 | 61.704 | 73.22 | 31.399 | 11.547 | 29.154 | 76.801 | 14.864 |
| 21 | 3.903 | 17.478 | 23.209 | 12.017 | 28.552 | 35.222 | 31.799 | 27.658 | 49.782 | 45.343 | 101.689 | 34.934 | 8.929 | 21.998 | 25.863 | 17.624 |
| 22 | 11.729 | 26.6 | 44.916 | 7.921 | 27.885 | 38.357 | 28.287 | 25.513 | 69.022 | 50.925 | 43.741 | 18.873 | 7.975 | 29.528 | 33.549 | 21.18 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.224 | -0.339 | 0.068 | -0.345 | 0.397 | 0.034 | 0.6 | -0.43 | -0.089 | 0.448 | 0.959 | -0.506 | -0.996 | 0.061 | -0.115 | -1.589 |
| 02 | -0.426 | 0.177 | 0.047 | -1.765 | -0.068 | 0.595 | -0.1 | -0.324 | 0.441 | 1.024 | 0.332 | -0.413 | -1.024 | 0.05 | -0.588 | -1.61 |
| 03 | 0.111 | -0.197 | 0.324 | -1.72 | 0.602 | 1.015 | 0.473 | -0.427 | -0.633 | 0.383 | 0.433 | -1.011 | -1.588 | -0.639 | -0.197 | -1.598 |
| 04 | -0.26 | -0.048 | -0.077 | 0.03 | -0.0 | 0.763 | 0.54 | -0.337 | -0.95 | 0.757 | 0.709 | -0.223 | -3.107 | -0.655 | -0.417 | -1.746 |
| 05 | -1.239 | 0.037 | -0.187 | -3.086 | 0.119 | 0.793 | 0.578 | -0.461 | 0.378 | 0.554 | 0.606 | -1.207 | -2.268 | -0.124 | 0.308 | -0.992 |
| 06 | -0.349 | -0.487 | 0.349 | -1.423 | 0.191 | 0.322 | 0.486 | 0.503 | -0.213 | 0.716 | 0.899 | -0.704 | -2.24 | -0.426 | -0.885 | -1.749 |
| 07 | 0.209 | -0.216 | -0.331 | -2.589 | 0.352 | -0.426 | 0.627 | -0.292 | 0.885 | -0.492 | 0.957 | -1.296 | -0.467 | -0.569 | 0.01 | -1.022 |
| 08 | -0.102 | 1.416 | -0.709 | -1.681 | -0.832 | 0.7 | -2.593 | -2.175 | -1.044 | 1.506 | -0.138 | -0.748 | -4.377 | 0.204 | -1.993 | -2.533 |
| 09 | -0.254 | -0.422 | -4.377 | -1.351 | 1.064 | 2.176 | -2.498 | -2.579 | -0.938 | 0.146 | -4.377 | -4.377 | -0.864 | -2.165 | -3.062 | -1.313 |
| 10 | -2.228 | -3.003 | 1.463 | -4.377 | -3.074 | -3.118 | 2.365 | -4.377 | -4.377 | -4.377 | -2.15 | -4.377 | -4.377 | -4.377 | -0.547 | -4.377 |
| 11 | -4.377 | -4.377 | -1.956 | -4.377 | -4.377 | -3.118 | -3.003 | -4.377 | -3.159 | -4.377 | 2.746 | -4.377 | -4.377 | -4.377 | -4.377 | -4.377 |
| 12 | -3.159 | -4.377 | -4.377 | -4.377 | -3.118 | -4.377 | -4.377 | -4.377 | 2.708 | -0.328 | -2.979 | -4.377 | -4.377 | -4.377 | -4.377 | -4.377 |
| 13 | 2.673 | -2.249 | -2.228 | -0.911 | -0.484 | -2.963 | -3.08 | -3.121 | -2.979 | -4.377 | -4.377 | -4.377 | -4.377 | -4.377 | -4.377 | -4.377 |
| 14 | 1.069 | -0.848 | 2.403 | -0.253 | -4.377 | -4.377 | -1.933 | -4.377 | -3.017 | -4.377 | -2.256 | -4.377 | -3.083 | -4.377 | -0.915 | -4.377 |
| 15 | -3.096 | 0.476 | -1.751 | 0.173 | -2.209 | -2.52 | -4.377 | -1.336 | 0.035 | 1.265 | -0.032 | 1.817 | -4.377 | -2.182 | -0.815 | -1.401 |
| 16 | -1.755 | -1.577 | -0.209 | -4.377 | -0.168 | -0.0 | 1.002 | -0.253 | -0.595 | -4.377 | -0.043 | -2.586 | -0.57 | 0.039 | 1.712 | -0.348 |
| 17 | -0.359 | -1.222 | 0.03 | -2.193 | -0.321 | -0.609 | -0.2 | -1.764 | 1.211 | 0.823 | 1.377 | -0.826 | -0.876 | -0.63 | -0.718 | -2.012 |
| 18 | 0.443 | 0.826 | 0.142 | -1.591 | -0.647 | 0.317 | 0.219 | -0.678 | 0.073 | 1.061 | 0.675 | -0.995 | -1.754 | -1.076 | -1.116 | -4.377 |
| 19 | -0.465 | -2.576 | 0.306 | 0.237 | -0.468 | 0.262 | 1.063 | 0.727 | -0.045 | 0.304 | 0.451 | -0.213 | -4.377 | -0.846 | -0.814 | -1.566 |
| 20 | -1.391 | -1.439 | 0.369 | -1.238 | -0.835 | -0.174 | 0.212 | -0.43 | -0.199 | 0.702 | 0.872 | 0.033 | -0.947 | -0.041 | 0.92 | -0.702 |
| 21 | -1.97 | -0.543 | -0.265 | -0.908 | -0.061 | 0.146 | 0.045 | -0.092 | 0.489 | 0.396 | 1.199 | 0.138 | -1.195 | -0.318 | -0.159 | -0.535 |
| 22 | -0.932 | -0.131 | 0.387 | -1.309 | -0.084 | 0.231 | -0.07 | -0.172 | 0.814 | 0.512 | 0.361 | -0.468 | -1.303 | -0.028 | 0.098 | -0.355 |