Model info
| Transcription factor | CTCFL | ||||||||
| Model | CTCFL_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 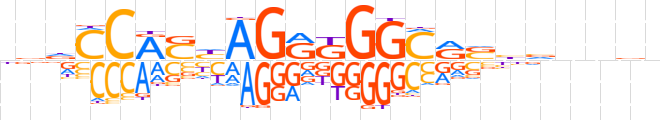 | ||||||||
| LOGO (reverse complement) | 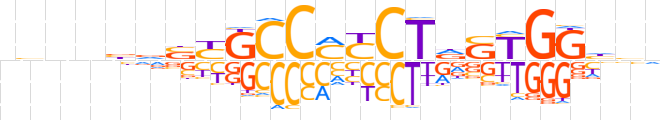 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 23 | ||||||||
| Quality | A | ||||||||
| Consensus | bbdCCRShAGRKGGCRSbvnnvn | ||||||||
| wAUC | 0.9162206759130554 | ||||||||
| Best AUC | 0.9369037874187588 | ||||||||
| Benchmark datasets | 3 | ||||||||
| Aligned words | 498 | ||||||||
| TF family | More than 3 adjacent zinc finger factors{2.3.3} | ||||||||
| TF subfamily | CTCF-like factors{2.3.3.50} | ||||||||
| HGNC | 16234 | ||||||||
| EntrezGene | 140690 | ||||||||
| UniProt ID | CTCFL_HUMAN | ||||||||
| UniProt AC | Q8NI51 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 5.725 | 11.122 | 24.456 | 11.046 | 42.223 | 39.245 | 35.956 | 59.923 | 10.536 | 16.847 | 24.674 | 8.726 | 18.553 | 40.566 | 117.448 | 29.038 |
| 02 | 26.732 | 3.066 | 38.514 | 8.724 | 49.537 | 11.72 | 24.029 | 22.494 | 47.509 | 21.313 | 105.349 | 28.361 | 22.312 | 10.857 | 51.804 | 23.76 |
| 03 | 11.1 | 123.963 | 6.824 | 4.203 | 3.879 | 39.205 | 1.003 | 2.869 | 17.29 | 179.979 | 7.88 | 14.548 | 1.817 | 74.627 | 3.019 | 3.878 |
| 04 | 0.864 | 29.445 | 2.863 | 0.913 | 4.159 | 411.601 | 0.969 | 1.045 | 0.983 | 14.751 | 2.991 | 0.0 | 1.989 | 21.554 | 0.0 | 1.954 |
| 05 | 0.864 | 0.864 | 2.005 | 4.262 | 379.543 | 6.698 | 51.884 | 39.227 | 3.038 | 0.904 | 1.912 | 0.969 | 0.913 | 0.0 | 1.045 | 1.954 |
| 06 | 3.874 | 241.514 | 130.817 | 8.153 | 2.737 | 5.729 | 0.0 | 0.0 | 3.022 | 35.521 | 17.428 | 0.875 | 0.983 | 21.762 | 19.836 | 3.831 |
| 07 | 0.0 | 6.765 | 1.016 | 2.835 | 43.936 | 127.481 | 8.931 | 124.177 | 13.837 | 109.097 | 13.799 | 31.349 | 0.983 | 3.091 | 5.79 | 2.995 |
| 08 | 43.646 | 5.403 | 4.058 | 5.65 | 235.061 | 0.0 | 7.498 | 3.875 | 24.956 | 0.0 | 3.583 | 0.997 | 144.739 | 0.894 | 12.652 | 3.07 |
| 09 | 1.915 | 4.854 | 441.632 | 0.0 | 0.904 | 0.0 | 5.393 | 0.0 | 1.873 | 0.0 | 23.843 | 2.074 | 0.0 | 2.011 | 11.582 | 0.0 |
| 10 | 0.969 | 0.0 | 2.746 | 0.976 | 3.808 | 0.0 | 2.03 | 1.028 | 179.213 | 2.927 | 295.472 | 4.838 | 0.0 | 0.0 | 1.003 | 1.071 |
| 11 | 4.903 | 0.0 | 124.747 | 54.341 | 0.0 | 0.983 | 0.0 | 1.944 | 7.032 | 11.561 | 165.35 | 117.308 | 0.0 | 0.0 | 6.885 | 1.028 |
| 12 | 2.012 | 0.0 | 8.96 | 0.962 | 1.833 | 0.0 | 8.868 | 1.844 | 2.013 | 0.904 | 294.066 | 0.0 | 0.0 | 0.0 | 174.621 | 0.0 |
| 13 | 0.0 | 0.0 | 4.889 | 0.969 | 0.904 | 0.0 | 0.0 | 0.0 | 3.85 | 10.138 | 436.473 | 36.054 | 0.0 | 0.922 | 1.884 | 0.0 |
| 14 | 0.0 | 3.8 | 0.0 | 0.954 | 2.976 | 5.151 | 1.929 | 1.003 | 42.3 | 368.271 | 15.587 | 17.087 | 0.0 | 31.743 | 5.28 | 0.0 |
| 15 | 7.092 | 2.961 | 32.452 | 2.771 | 190.706 | 9.603 | 196.755 | 11.901 | 5.869 | 1.929 | 10.788 | 4.211 | 0.0 | 1.003 | 18.041 | 0.0 |
| 16 | 10.537 | 56.56 | 131.281 | 5.29 | 1.905 | 9.8 | 0.0 | 3.791 | 26.247 | 187.933 | 29.312 | 14.544 | 2.888 | 7.021 | 6.975 | 1.998 |
| 17 | 6.949 | 13.977 | 4.009 | 16.641 | 34.129 | 87.859 | 16.464 | 122.862 | 18.731 | 57.503 | 33.426 | 57.908 | 1.885 | 7.903 | 8.759 | 7.077 |
| 18 | 21.326 | 14.893 | 19.427 | 6.048 | 94.549 | 35.484 | 19.786 | 17.423 | 9.925 | 29.153 | 20.084 | 3.497 | 48.182 | 45.577 | 101.037 | 9.691 |
| 19 | 30.528 | 35.707 | 74.998 | 32.749 | 34.383 | 20.986 | 24.888 | 44.851 | 19.693 | 41.511 | 22.658 | 76.473 | 7.758 | 12.051 | 11.836 | 5.013 |
| 20 | 29.115 | 23.448 | 25.878 | 13.92 | 35.244 | 31.573 | 11.087 | 32.351 | 49.373 | 33.066 | 23.534 | 28.408 | 20.202 | 26.394 | 55.721 | 56.768 |
| 21 | 28.627 | 21.217 | 71.732 | 12.356 | 45.357 | 17.842 | 27.19 | 24.093 | 27.071 | 35.205 | 37.959 | 15.985 | 14.706 | 26.135 | 70.982 | 19.625 |
| 22 | 15.6 | 36.973 | 34.542 | 28.645 | 35.317 | 33.918 | 15.771 | 15.393 | 66.402 | 59.25 | 50.024 | 32.189 | 8.851 | 25.184 | 24.449 | 13.577 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | -1.636 | -1.003 | -0.234 | -1.01 | 0.306 | 0.233 | 0.146 | 0.653 | -1.056 | -0.6 | -0.225 | -1.237 | -0.505 | 0.266 | 1.323 | -0.065 |
| 02 | -0.146 | -2.207 | 0.214 | -1.237 | 0.464 | -0.953 | -0.251 | -0.316 | 0.422 | -0.369 | 1.214 | -0.088 | -0.324 | -1.027 | 0.508 | -0.262 |
| 03 | -1.005 | 1.377 | -1.471 | -1.923 | -1.996 | 0.232 | -3.116 | -2.266 | -0.574 | 1.748 | -1.334 | -0.743 | -2.656 | 0.871 | -2.221 | -1.996 |
| 04 | -3.222 | -0.051 | -2.268 | -3.184 | -1.932 | 2.574 | -3.141 | -3.087 | -3.131 | -0.729 | -2.229 | -4.394 | -2.581 | -0.358 | -4.394 | -2.595 |
| 05 | -3.222 | -3.222 | -2.574 | -1.91 | 2.493 | -1.488 | 0.51 | 0.233 | -2.215 | -3.191 | -2.614 | -3.141 | -3.184 | -4.394 | -3.087 | -2.595 |
| 06 | -1.997 | 2.042 | 1.43 | -1.302 | -2.307 | -1.636 | -4.394 | -4.394 | -2.22 | 0.134 | -0.566 | -3.213 | -3.131 | -0.349 | -0.44 | -2.007 |
| 07 | -4.394 | -1.479 | -3.107 | -2.276 | 0.345 | 1.404 | -1.215 | 1.378 | -0.792 | 1.249 | -0.794 | 0.011 | -3.131 | -2.2 | -1.626 | -2.228 |
| 08 | 0.338 | -1.69 | -1.955 | -1.648 | 2.015 | -4.394 | -1.382 | -1.997 | -0.214 | -4.394 | -2.068 | -3.121 | 1.531 | -3.198 | -0.879 | -2.206 |
| 09 | -2.612 | -1.79 | 2.645 | -4.394 | -3.191 | -4.394 | -1.692 | -4.394 | -2.631 | -4.394 | -0.259 | -2.546 | -4.394 | -2.571 | -0.964 | -4.394 |
| 10 | -3.141 | -4.394 | -2.304 | -3.136 | -2.012 | -4.394 | -2.564 | -3.099 | 1.744 | -2.248 | 2.243 | -1.793 | -4.394 | -4.394 | -3.116 | -3.069 |
| 11 | -1.781 | -4.394 | 1.383 | 0.556 | -4.394 | -3.131 | -4.394 | -2.6 | -1.442 | -0.966 | 1.664 | 1.322 | -4.394 | -4.394 | -1.462 | -3.099 |
| 12 | -2.571 | -4.394 | -1.211 | -3.147 | -2.648 | -4.394 | -1.221 | -2.644 | -2.571 | -3.191 | 2.239 | -4.394 | -4.394 | -4.394 | 1.718 | -4.394 |
| 13 | -4.394 | -4.394 | -1.783 | -3.141 | -3.191 | -4.394 | -4.394 | -4.394 | -2.003 | -1.093 | 2.633 | 0.149 | -4.394 | -3.177 | -2.626 | -4.394 |
| 14 | -4.394 | -2.014 | -4.394 | -3.152 | -2.233 | -1.735 | -2.606 | -3.116 | 0.307 | 2.463 | -0.676 | -0.586 | -4.394 | 0.023 | -1.712 | -4.394 |
| 15 | -1.434 | -2.238 | 0.045 | -2.296 | 1.806 | -1.145 | 1.837 | -0.938 | -1.613 | -2.606 | -1.033 | -1.921 | -4.394 | -3.116 | -0.533 | -4.394 |
| 16 | -1.056 | 0.596 | 1.434 | -1.71 | -2.617 | -1.125 | -4.394 | -2.016 | -0.164 | 1.792 | -0.055 | -0.743 | -2.26 | -1.444 | -1.45 | -2.577 |
| 17 | -1.454 | -0.782 | -1.966 | -0.612 | 0.095 | 1.034 | -0.622 | 1.368 | -0.496 | 0.612 | 0.074 | 0.619 | -2.625 | -1.331 | -1.233 | -1.436 |
| 18 | -0.369 | -0.72 | -0.46 | -1.585 | 1.107 | 0.133 | -0.442 | -0.567 | -1.113 | -0.061 | -0.428 | -2.09 | 0.436 | 0.381 | 1.173 | -1.136 |
| 19 | -0.015 | 0.14 | 0.876 | 0.054 | 0.102 | -0.384 | -0.217 | 0.365 | -0.447 | 0.289 | -0.309 | 0.895 | -1.349 | -0.926 | -0.943 | -1.76 |
| 20 | -0.062 | -0.275 | -0.178 | -0.786 | 0.127 | 0.018 | -1.006 | 0.042 | 0.461 | 0.064 | -0.272 | -0.086 | -0.422 | -0.159 | 0.581 | 0.599 |
| 21 | -0.079 | -0.374 | 0.832 | -0.902 | 0.376 | -0.544 | -0.13 | -0.249 | -0.134 | 0.126 | 0.2 | -0.651 | -0.732 | -0.169 | 0.821 | -0.45 |
| 22 | -0.675 | 0.174 | 0.107 | -0.078 | 0.129 | 0.089 | -0.664 | -0.688 | 0.755 | 0.642 | 0.474 | 0.037 | -1.223 | -0.205 | -0.234 | -0.81 |