Model info
| Transcription factor | Crx | ||||||||
| Model | CRX_MOUSE.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 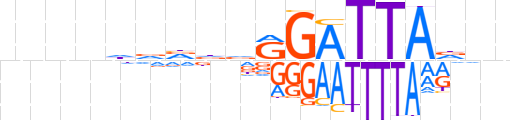 | ||||||||
| LOGO (reverse complement) | 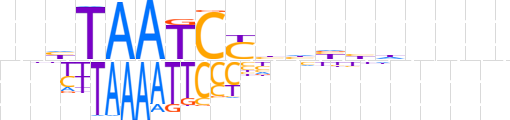 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 18 | ||||||||
| Quality | A | ||||||||
| Consensus | nnnnhdvddRGATTAdnn | ||||||||
| wAUC | 0.862689719482612 | ||||||||
| Best AUC | 0.8724564705431574 | ||||||||
| Benchmark datasets | 2 | ||||||||
| Aligned words | 500 | ||||||||
| TF family | Paired-related HD factors{3.1.3} | ||||||||
| TF subfamily | OTX{3.1.3.17} | ||||||||
| MGI | 1194883 | ||||||||
| EntrezGene | 12951 | ||||||||
| UniProt ID | CRX_MOUSE | ||||||||
| UniProt AC | O54751 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 43.858 | 7.883 | 61.722 | 32.118 | 28.961 | 10.913 | 1.743 | 42.519 | 57.176 | 23.88 | 39.701 | 15.126 | 37.088 | 27.863 | 30.969 | 38.479 |
| 02 | 34.795 | 34.576 | 45.727 | 51.985 | 16.57 | 8.987 | 0.0 | 44.982 | 40.281 | 36.309 | 44.659 | 12.887 | 24.9 | 25.623 | 30.168 | 47.55 |
| 03 | 26.278 | 25.349 | 35.603 | 29.317 | 25.157 | 19.288 | 2.943 | 58.107 | 29.279 | 52.776 | 17.437 | 21.063 | 31.777 | 36.33 | 23.591 | 65.706 |
| 04 | 46.482 | 4.055 | 20.242 | 41.712 | 36.063 | 21.427 | 0.925 | 75.329 | 21.783 | 14.376 | 15.146 | 28.269 | 51.288 | 29.56 | 25.204 | 68.14 |
| 05 | 76.301 | 12.792 | 46.632 | 19.891 | 37.141 | 9.211 | 1.825 | 21.24 | 16.029 | 9.428 | 27.062 | 8.998 | 91.857 | 20.021 | 63.286 | 38.285 |
| 06 | 146.516 | 6.775 | 57.414 | 10.623 | 33.486 | 5.088 | 1.77 | 11.108 | 56.517 | 27.971 | 36.85 | 17.468 | 36.311 | 19.221 | 20.485 | 12.398 |
| 07 | 44.591 | 15.349 | 140.933 | 71.957 | 26.213 | 2.943 | 1.876 | 28.023 | 48.115 | 14.666 | 37.254 | 16.485 | 7.804 | 9.81 | 28.339 | 5.643 |
| 08 | 39.702 | 8.721 | 66.145 | 12.155 | 20.28 | 6.151 | 2.967 | 13.369 | 80.189 | 40.75 | 59.556 | 27.907 | 33.299 | 15.792 | 54.943 | 18.073 |
| 09 | 32.971 | 0.0 | 139.421 | 1.078 | 38.9 | 4.02 | 23.779 | 4.715 | 55.786 | 3.72 | 115.124 | 8.98 | 16.934 | 0.848 | 52.847 | 0.876 |
| 10 | 0.0 | 0.0 | 144.591 | 0.0 | 0.0 | 1.75 | 6.838 | 0.0 | 8.909 | 15.871 | 306.391 | 0.0 | 0.0 | 0.0 | 15.65 | 0.0 |
| 11 | 6.944 | 1.965 | 0.0 | 0.0 | 16.735 | 0.885 | 0.0 | 0.0 | 396.266 | 74.475 | 1.758 | 0.973 | 0.0 | 0.0 | 0.0 | 0.0 |
| 12 | 0.0 | 0.0 | 0.0 | 419.945 | 0.0 | 0.0 | 0.0 | 77.324 | 0.0 | 0.0 | 0.0 | 1.758 | 0.0 | 0.0 | 0.0 | 0.973 |
| 13 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.872 | 3.106 | 0.0 | 495.021 |
| 14 | 1.872 | 0.0 | 0.0 | 0.0 | 3.106 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 479.684 | 0.0 | 3.737 | 11.6 |
| 15 | 221.815 | 31.23 | 171.858 | 59.76 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 1.898 | 0.0 | 1.838 | 2.96 | 2.092 | 6.548 | 0.0 |
| 16 | 77.573 | 50.754 | 73.907 | 22.541 | 18.775 | 9.993 | 0.0 | 6.453 | 37.739 | 52.507 | 58.107 | 30.052 | 9.049 | 20.188 | 27.182 | 5.178 |
| 17 | 30.027 | 18.213 | 70.415 | 24.482 | 26.056 | 52.432 | 2.961 | 51.993 | 35.41 | 42.579 | 43.849 | 37.359 | 13.223 | 15.664 | 21.442 | 13.895 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.335 | -1.342 | 0.675 | 0.027 | -0.075 | -1.029 | -2.697 | 0.305 | 0.599 | -0.265 | 0.237 | -0.713 | 0.169 | -0.113 | -0.009 | 0.206 |
| 02 | 0.106 | 0.1 | 0.377 | 0.504 | -0.624 | -1.216 | -4.4 | 0.36 | 0.251 | 0.148 | 0.353 | -0.868 | -0.224 | -0.196 | -0.035 | 0.416 |
| 03 | -0.171 | -0.206 | 0.129 | -0.063 | -0.214 | -0.475 | -2.251 | 0.615 | -0.064 | 0.519 | -0.574 | -0.389 | 0.017 | 0.149 | -0.277 | 0.737 |
| 04 | 0.393 | -1.963 | -0.428 | 0.286 | 0.142 | -0.372 | -3.182 | 0.873 | -0.356 | -0.762 | -0.711 | -0.099 | 0.491 | -0.055 | -0.212 | 0.773 |
| 05 | 0.885 | -0.876 | 0.396 | -0.445 | 0.171 | -1.193 | -2.66 | -0.38 | -0.656 | -1.17 | -0.142 | -1.215 | 1.07 | -0.438 | 0.699 | 0.201 |
| 06 | 1.535 | -1.485 | 0.603 | -1.055 | 0.068 | -1.754 | -2.685 | -1.012 | 0.587 | -0.109 | 0.163 | -0.572 | 0.148 | -0.478 | -0.416 | -0.906 |
| 07 | 0.352 | -0.698 | 1.497 | 0.827 | -0.173 | -2.251 | -2.637 | -0.108 | 0.427 | -0.743 | 0.174 | -0.629 | -1.351 | -1.132 | -0.097 | -1.657 |
| 08 | 0.237 | -1.245 | 0.743 | -0.925 | -0.426 | -1.576 | -2.244 | -0.833 | 0.935 | 0.263 | 0.639 | -0.112 | 0.063 | -0.671 | 0.559 | -0.539 |
| 09 | 0.053 | -4.4 | 1.486 | -3.071 | 0.217 | -1.971 | -0.269 | -1.824 | 0.574 | -2.041 | 1.295 | -1.217 | -0.602 | -3.242 | 0.52 | -3.22 |
| 10 | -4.4 | -4.4 | 1.522 | -4.4 | -4.4 | -2.694 | -1.477 | -4.4 | -1.225 | -0.666 | 2.272 | -4.4 | -4.4 | -4.4 | -0.679 | -4.4 |
| 11 | -1.462 | -2.599 | -4.4 | -4.4 | -0.614 | -3.213 | -4.4 | -4.4 | 2.529 | 0.861 | -2.691 | -3.146 | -4.4 | -4.4 | -4.4 | -4.4 |
| 12 | -4.4 | -4.4 | -4.4 | 2.587 | -4.4 | -4.4 | -4.4 | 0.899 | -4.4 | -4.4 | -4.4 | -2.691 | -4.4 | -4.4 | -4.4 | -3.146 |
| 13 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -2.639 | -2.203 | -4.4 | 2.751 |
| 14 | -2.639 | -4.4 | -4.4 | -4.4 | -2.203 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | 2.72 | -4.4 | -2.037 | -0.97 |
| 15 | 1.949 | -0.001 | 1.695 | 0.642 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -2.627 | -4.4 | -2.654 | -2.246 | -2.546 | -1.518 | -4.4 |
| 16 | 0.902 | 0.48 | 0.854 | -0.322 | -0.501 | -1.114 | -4.4 | -1.531 | 0.187 | 0.514 | 0.615 | -0.039 | -1.21 | -0.43 | -0.138 | -1.738 |
| 17 | -0.039 | -0.531 | 0.806 | -0.241 | -0.179 | 0.513 | -2.246 | 0.504 | 0.124 | 0.306 | 0.335 | 0.177 | -0.843 | -0.678 | -0.371 | -0.795 |