Model info
| Transcription factor | EBF1 | ||||||||
| Model | COE1_HUMAN.H10DI.A | ||||||||
| Model type | Dinucleotide PWM | ||||||||
| LOGO | 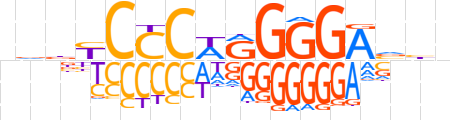 | ||||||||
| LOGO (reverse complement) | 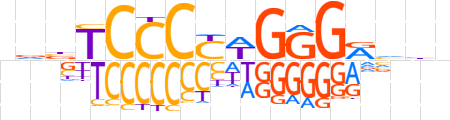 | ||||||||
| Origin | ChIP-Seq | ||||||||
| Model length | 16 | ||||||||
| Quality | A | ||||||||
| Consensus | ndbYCCCWRGGGAvhn | ||||||||
| wAUC | 0.8608839163528562 | ||||||||
| Best AUC | 0.9249900857816359 | ||||||||
| Benchmark datasets | 3 | ||||||||
| Aligned words | 503 | ||||||||
| TF family | Early B-Cell Factor-related factors{6.1.5} | ||||||||
| TF subfamily | EBF1 (COE1){6.1.5.0.1} | ||||||||
| HGNC | 3126 | ||||||||
| EntrezGene | 1879 | ||||||||
| UniProt ID | COE1_HUMAN | ||||||||
| UniProt AC | Q9UH73 | ||||||||
| Comment | |||||||||
| Downloads | pcm
pwm alignment threshold to P-value grid | ||||||||
| Standard thresholds |
|
PCM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 63.179 | 13.68 | 45.608 | 23.808 | 64.711 | 27.414 | 15.074 | 34.299 | 32.656 | 11.282 | 36.752 | 18.418 | 31.691 | 6.915 | 51.601 | 22.913 |
| 02 | 24.316 | 30.374 | 80.022 | 57.525 | 13.625 | 25.521 | 1.858 | 18.287 | 6.928 | 30.831 | 82.139 | 29.136 | 4.764 | 21.789 | 40.524 | 32.361 |
| 03 | 0.965 | 8.962 | 2.668 | 37.037 | 4.078 | 31.76 | 0.0 | 72.676 | 6.746 | 73.469 | 16.883 | 107.445 | 7.458 | 48.388 | 6.032 | 75.431 |
| 04 | 0.0 | 19.247 | 0.0 | 0.0 | 0.974 | 161.606 | 0.0 | 0.0 | 0.0 | 25.584 | 0.0 | 0.0 | 0.0 | 290.73 | 1.859 | 0.0 |
| 05 | 0.0 | 0.974 | 0.0 | 0.0 | 5.723 | 385.701 | 0.0 | 105.743 | 0.0 | 1.859 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 06 | 2.041 | 3.682 | 0.0 | 0.0 | 3.14 | 377.61 | 0.0 | 7.785 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 102.381 | 1.682 | 1.68 |
| 07 | 1.009 | 0.0 | 2.081 | 2.09 | 256.767 | 6.914 | 7.656 | 212.335 | 0.837 | 0.0 | 0.0 | 0.845 | 1.68 | 0.0 | 1.895 | 5.89 |
| 08 | 84.929 | 7.733 | 146.373 | 21.258 | 0.0 | 0.0 | 6.914 | 0.0 | 1.967 | 0.0 | 9.666 | 0.0 | 32.152 | 0.0 | 189.008 | 0.0 |
| 09 | 0.0 | 0.0 | 119.048 | 0.0 | 1.069 | 0.0 | 6.664 | 0.0 | 5.224 | 0.0 | 345.755 | 0.983 | 0.0 | 0.0 | 21.258 | 0.0 |
| 10 | 0.905 | 0.0 | 4.397 | 0.99 | 0.0 | 0.0 | 0.0 | 0.0 | 60.092 | 0.933 | 431.7 | 0.0 | 0.0 | 0.0 | 0.0 | 0.983 |
| 11 | 1.084 | 1.622 | 58.292 | 0.0 | 0.0 | 0.933 | 0.0 | 0.0 | 0.0 | 4.635 | 430.63 | 0.833 | 0.0 | 0.983 | 0.99 | 0.0 |
| 12 | 1.084 | 0.0 | 0.0 | 0.0 | 6.437 | 0.0 | 1.734 | 0.0 | 405.653 | 1.93 | 69.92 | 12.409 | 0.833 | 0.0 | 0.0 | 0.0 |
| 13 | 79.028 | 179.346 | 130.353 | 25.28 | 0.974 | 0.956 | 0.0 | 0.0 | 20.686 | 30.667 | 16.657 | 3.644 | 1.645 | 3.342 | 7.422 | 0.0 |
| 14 | 23.227 | 22.652 | 2.632 | 53.821 | 56.247 | 71.935 | 17.294 | 68.835 | 28.074 | 67.046 | 20.969 | 38.342 | 1.953 | 6.76 | 3.559 | 16.652 |
| 15 | 27.541 | 22.556 | 34.181 | 25.223 | 45.478 | 45.324 | 8.018 | 69.574 | 11.815 | 11.584 | 14.524 | 6.532 | 32.809 | 37.789 | 48.436 | 58.616 |
PWM
| AA | AC | AG | AT | CA | CC | CG | CT | GA | GC | GG | GT | TA | TC | TG | TT | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 0.698 | -0.81 | 0.374 | -0.268 | 0.722 | -0.129 | -0.716 | 0.092 | 0.043 | -0.997 | 0.16 | -0.52 | 0.014 | -1.466 | 0.497 | -0.306 |
| 02 | -0.247 | -0.028 | 0.933 | 0.605 | -0.814 | -0.2 | -2.645 | -0.527 | -1.464 | -0.013 | 0.959 | -0.069 | -1.815 | -0.355 | 0.257 | 0.035 |
| 03 | -3.152 | -1.219 | -2.337 | 0.168 | -1.958 | 0.016 | -4.4 | 0.837 | -1.489 | 0.848 | -0.605 | 1.226 | -1.394 | 0.433 | -1.595 | 0.874 |
| 04 | -4.4 | -0.477 | -4.4 | -4.4 | -3.145 | 1.633 | -4.4 | -4.4 | -4.4 | -0.197 | -4.4 | -4.4 | -4.4 | 2.219 | -2.644 | -4.4 |
| 05 | -4.4 | -3.145 | -4.4 | -4.4 | -1.644 | 2.502 | -4.4 | 1.21 | -4.4 | -2.644 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 |
| 06 | -2.567 | -2.051 | -4.4 | -4.4 | -2.194 | 2.481 | -4.4 | -1.353 | -4.4 | -4.4 | -4.4 | -4.4 | -4.4 | 1.178 | -2.726 | -2.728 |
| 07 | -3.12 | -4.4 | -2.55 | -2.547 | 2.095 | -1.466 | -1.369 | 1.906 | -3.251 | -4.4 | -4.4 | -3.245 | -2.728 | -4.4 | -2.629 | -1.617 |
| 08 | 0.992 | -1.36 | 1.534 | -0.38 | -4.4 | -4.4 | -1.466 | -4.4 | -2.598 | -4.4 | -1.146 | -4.4 | 0.028 | -4.4 | 1.789 | -4.4 |
| 09 | -4.4 | -4.4 | 1.328 | -4.4 | -3.078 | -4.4 | -1.501 | -4.4 | -1.729 | -4.4 | 2.392 | -3.139 | -4.4 | -4.4 | -0.38 | -4.4 |
| 10 | -3.197 | -4.4 | -1.889 | -3.134 | -4.4 | -4.4 | -4.4 | -4.4 | 0.648 | -3.176 | 2.614 | -4.4 | -4.4 | -4.4 | -4.4 | -3.139 |
| 11 | -3.068 | -2.756 | 0.618 | -4.4 | -4.4 | -3.176 | -4.4 | -4.4 | -4.4 | -1.84 | 2.612 | -3.255 | -4.4 | -3.139 | -3.134 | -4.4 |
| 12 | -3.068 | -4.4 | -4.4 | -4.4 | -1.534 | -4.4 | -2.702 | -4.4 | 2.552 | -2.614 | 0.799 | -0.905 | -3.255 | -4.4 | -4.4 | -4.4 |
| 13 | 0.92 | 1.737 | 1.419 | -0.209 | -3.145 | -3.158 | -4.4 | -4.4 | -0.406 | -0.019 | -0.618 | -2.06 | -2.745 | -2.138 | -1.399 | -4.4 |
| 14 | -0.292 | -0.317 | -2.349 | 0.538 | 0.582 | 0.827 | -0.582 | 0.783 | -0.106 | 0.757 | -0.393 | 0.202 | -2.604 | -1.487 | -2.081 | -0.619 |
| 15 | -0.125 | -0.321 | 0.089 | -0.211 | 0.371 | 0.368 | -1.325 | 0.794 | -0.953 | -0.972 | -0.752 | -1.52 | 0.048 | 0.188 | 0.434 | 0.623 |